INTRODUCTION
Swiprosin-1 was first identified in human lymphocytes, predominantly in CD8+ lymphocytes (1) and later in immature B cells, resting and activated B cells, and non-lymphoid tissue, especially in the brain (2). We have recently found that swiprosin-1 is also expressed in mast cells and up-regulated in both in vitro cultured mast cells by phorbol ester or cross-linking of FcεR1 and in vivo model tissues of passive cutaneous anaphylaxis and atopic dermatitis (3). Targeted inhibition of the specific protein kinase C (PKC) isotypes by siRNA revealed that PKC-βI/η are involved in the expression of swiprosin-1 in the human mast cell line HMC-1. In contrast, down-regulation of swiprosin-1 by A23187 or ionomycin suggests that calcium-signaling plays a negative role (3).
However, the functions of swiprosin-1 in mast cells are still largely unknown. In the previous paper, we only reported that the ectopic expression of swiprosin-1 augments PMA/A23187-induced NF-κB promoter activity and cytokine expression including IL-3 and IL-8 (3). However, the mechanism how swiprosin-1 involves in the cytokine production in mast cells is not investigated. In other cell types, the only reported functions are that swiprosin-1 is associated with lipid rafts in the immature B-cell line WEHI231 and that it participates in enhancement of BCR signals and contributes to BCR-induced apoptosis (2,4).
Mast cells are broadly distributed throughout mammalian tissues and play a critical role in a variety of biological responses (5-7). Typically, mast cells are considered in association with immediate-type hypersensitivity (5). However, several recent reports have provided evidence for the possible participation of mast cells in more persistent, and even in chronic, inflammatory and immunological, responses (8,9). Of note, a variety of cytokines including IL-3, IL-4, IL-5, IL-6, IL-8, TNF-α, and IFN-γ (10-12) are produced in mast cells and play an important role in immunological processes other than IgE-mediated hypersensitivity reactions. Given the potential importance of mast cell-derived cytokines in physiological or pathological immune reactions, it is essential to understand the signaling pathways and molecules involved in cytokine regulation in mast cells. Until recently, however, only a limited number of reports have examined the regulatory mechanism of cytokine expression in mast cells, while the mechanism of mast cell degranulation, mediated by the high affinity IgE receptor (FcεR1), is relatively well characterized (7).
As part of genome-wide approaches to finding novel genes that may be involved in mast cell activation, we have previously found that swiprosin-1 is over-induced in the human mast cell line HMC-1 stimulated with PMA/A23187 (3). In the present study, we examined, using confocal microscopy, the three-dimensional localization of swiprosin-1 in various cell lines, including HMC-1 cells, 293T cells, and COS-7 cells. We then asked whether swiprosin-1 potentially modulates mast cell activation and cytokine expression in relation to its localization.
Database mining revealed that swiprosin-1 putatively contains four myristylation sites, three binding sites for SH3 domain containing proteins, two potential EF-hand domains, and a coiled-coil domain at the C-terminus, and therefore, may have a role as a small adaptor protein involved in calcium signaling (1). In accordance with this prediction, swiprosin- 1 was implicated in phosphotyrosine-based signaling events involved in the cellular stimulation of early growth factor (EGF)3 and in actin rearrangement (13). By utilizing HMC-1 cell line, which was established from a patient with mast cell leukemia, we studied whether swiprosin-1 involves in the expression of human cytokines and chemokines. The results presented here strongly demonstrate that swiprosin-1 potentially acts as a regulator for cytokine expression and activation of mast cells.
MATERIALS AND METHODS
Antibodies and reagents
Goat polyclonal antibody to swiprosin-1 was from Imgenex (San Diego, CA). Antibodies to p-PI3K, p-Akt, and GFP were from Cell Signaling Technology, Inc (Beverly, MA). HRP-conjugated anti-goat, anti-rabbit, and anti-mouse IgGs were from GE Healthcare (Chalfont St. Giles, United Kingdom). SB-203580, PD98059, MG132, cyclosporine A, and PP2 were purchased from Calbiochem-Behring (La Jolla, CA). Total RNA isolation reagent was from WelPrep™ Join Bio Innovation (Daegu, South Korea). Maxime RT Premix (oligo dT primer), Maxime PCR PreMix, and a plasmid purification kit were from iNtRON Biotechnology (Daejon, South Korea). SYBR premix Ex Taq was from Takara Bio Inc (Shiga, Japan). The dual-luciferase reporter assay system was from Promega Corporation (Madison, WI). The ELISA kit for hIL-8 was from R&D Systems (Minneapolis, MN). All other reagents used in this study were purchased from Sigma Chemical Co (St. Louis, MO).
Cell culture
HMC-1 cells were cultured in IMDM medium. Jurkat T and Molt-4 T cells (T cells), Raji B cells (B cells), THP-1 cells (monocytes), and 293-T and CHO-K1 cells (epithelial cells) were maintained in RPMI 1640 medium. J774A.1 cells (macrophages), HT-29 cells (epithelial cells), COS-7 cells (fibroblasts), and RBL-2H3 cells (mast cells) were cultured in DMEM medium. HUVECs were cultured in EBM medium. All culture media used in this study were supplemented with 10% heat inactivated FBS, 100 units/ml penicillin G, and 100µg/ml streptomycin.
Recombinant DNA constructs
To generate the swiprosin-1/pEGFP-C1 construct, the human swiprosin-1 clone coding for the full-length open reading frame of swiprosin-1 in the pOTB7 vector was purchased from RZPD German Resource Center (Berlin, Germany). This was used as a template, and a PCR amplification was performed using the primers: sense 5'-AAGAATTCTATGGCCACGGACGAGCTGGCCACC-3' containing the EcoRI restriction site and anti-sense 5'-TTTGGATCCCTACTTAAAGGTGGACTGCAGCTC-3' containing the Bam HI restriction site. The PCR product was subcloned as a EcoRI/Bam HI fragment into a pEGFP-C1 vector (Clontech Laboratory, Inc.) of a neomycin resistant gene, resulting in an in-frame fusion of swiprosin-1 to the COOH terminus of GFP. The amino acid sequences of a linker polypeptide between EGFP and swiprosin-1 were SGLRSRAQAS (10 aa). To generate wild-type swiprosin-1 in a pcDNA3 vector or in a pHJ-1 vector (lentiviral vector), swiprosin-1 cDNA from the pOTB7 vector was transferred into the pcDNA3 vector by EcoRI/XhoI or the pHJ-1 vector by Not I restriction digestions and confirmed by automated sequencing.
Establishment of stable cell lines
Stable HMC-1 transfectants were established using the Nucleofector device and corresponding kits (Amaxa, Cologne, Germany) by introducing pEGFP-C1 or swiprosin-1/pEGFP-C1 cDNA followed by selection with 1 mg/ml geneticin (Invitrogen). The cells were then subjected to a FACS and sorted to selectively obtain GFP-positive cells. They were then cultured in complete medium supplemented with the same concentrations of antibiotics. The stable cells that expressed EGFP or EGFP fused with swiprosin-1 were designated as H-GFP cells and H-swip-1_GFP cells, respectively.
RNA isolation and RT-PCR
Cells from the tissue samples or in vitro cultures were harvested and total RNA was isolated using the WelPrep™ JBI method (iNtRON Biotechnology, Daejon, Korea) according to the manufacturer's instructions. Reverse transcription of the RNA was performed using oligo dT primer Maxime RT-PCR PreMix (iNtRON Biotechnology, Daejon, Korea). Two micrograms of RNA was transferred to an oligo dT primer mixture tube. The reaction volume was 20µl. cDNA synthesis was performed at 45℃ for 60 min, followed by RT inactivation at 95℃ for 5 min. Thereafter, the RT-generated DNA was diluted to 40µl volume with distilled water. The diluted RT-generated DNA (2µl) was amplified using Maxime PCR PreMix (iNtRON Biotechnology, Daejon, Korea). The primers used for cDNA amplification were as follows: hSwip-1(p1), sense 5'-ATCTTCCGCAAGGCGGCGGCCGGGGAG-3' and antisense 5'-GACTGCAGCTCCTTGAAGGCCGCTTTC-3'; hSwip-1 (p2), sense 5'-CTCGAGGCCACAGTTATGCAA-3' and antisense 5-'TAAGGCAAACGCA-3'; rSwip-1(p1), Sense 5'-CGTTATGCAATGTTCCTCGTGTCACTG-3' and antisense 5'-GATTCTCACAGGTTCTAAGACCGAAAA-3'; rSwip-1(p2), Sense 5'-TACAATGCCTCAAGAGCCCCCGGGACC-3' and antisense 5'-CCAAAAGTAAAGGGAATTTATATTCCT-3'; mSwip-1, sense 5'-GAGGCTCCCGACGAGACTGCCCAGGCG-3' and antisense 5'-CCGGAAGCTGAGTTTGCTGTCGAAATC-3'; hIL-3, sense 5'-CTTTGCCTTTGCTGGACTTC-3' and antisense 5'-CGAGGCTCAAAGTCGTCTG-3'; hIL-8, sense 5'-GTGCAGTTTTGCCAAGGAGT-3' and antisense 5'-CTCTGCACCCAGTTTTCCTT-3'; hTNF-α, sense 5'-GGCTCCAGGCGGTGCTTGTTC-3' and antisense 5'-AGACGGCGATGCGGCTGATG-3'; hGAPDH, sense 5'-CGGAGTCAACGGATTTGGTCGTAT-3' and antisense 5'-AGCCTTCTCCATGGTGGTGAAGAC-3'; rβ-actin, sense 5'-ATTGAACACGGCATTGTCAC-3' and antisense 5'-GTCTCAAACATGATCTGGGTC-3'. Amplification conditions were denaturation at 94℃ for 30 s, annealing at 58~68℃ for 20 s, and extension at 72℃ for 40 s for 30~35 cycles. The PCR products were resolved and visualized on a 1 or 1.5% agarose gel and stained with ethidium bromide.
Real-time quantitative RT-PCR
In all the experiments, the expression levels of the examined genes were evaluated by real-time RT-PCR, unless otherwise indicated. PCR amplification was performed in DNA Engine Opticon for a continuous fluorescence detection system (MJ Research, Waltham, MA) in a total volume of 20µl containing 2µl of cDNA/control and gene specific primers using the SYBR premix Ex Taq kit (Takara, Shiga, Japan). The PCR was performed under the following conditions: 94℃ for 30 s, 58~68℃ for 30 s, 72℃ for 30 s, plate read (detection of fluorescent product) for 40 cycles, followed by 7 min of extension at 72℃. A melting curve analysis was done to characterize the dsDNA product by slowly raising the temperature (0.2℃/s) from 65℃ to 95℃ with fluorescence data collected at 0.2℃ intervals. The levels of expression (of swiprosin-1, IL-3, IL-8, and TNF-α) that were normalized by GAPDH or β-actin were expressed as a relative value (i.e., percentage) of the maximum. The result of the maximum level in each experiment was considered as 100%.
Immunofluorescence staining and confocal imaging analysis
Stable HMC-1 cells (H-GFP or H-swip-1_GFP; 1×105) or COS-7 cells (transfected with GFP or swip-1_GFP) were seeded onto the poly-L-lysine (PLL)-coated or uncoated 18-mm glass coverslips. The cells were then fixed with 3.7% formaldehyde for 10 min and washed twice with 1X PBS. F-actin was detected by staining with phalloidin-TRITC (50µg/ml). 293-T cells (swiprosin-1 transfectants) were also grown on glass coverslips. The cells were fixed as described above, then incubated with primary goat polyclonal swiprosin-1 antibody, followed by secondary FITC-conjugated anti-goat antibody. The localization of swiprosin-1 and actin was examined with an FV1000 confocal laser scanning microscope (Olympus Corporation, Japan) equipped with 40×, 63×, and 100× objectives. In some cases, cells were not fixed and live images were obtained by confocal microscopy. For the co-localization analysis, the Z section cutting area through the apical surface was chosen. The overlapping degree of intensity (ODI) was calculated by FLUOVIEW software version 1.5. The co-localization percentage was then derived from the ODI, multiplying by 100.
For live-cell time-lapse confocal imaging, H-swip-1_GFP cells were seeded at 2×105 on 18-mm glass coverslips mounted in a chamber device. Cells were allowed to settle for 10 min at 37℃ and then activated by adding PMA/A23187. During the live-cell imaging, the chamber devices were maintained at 37℃ in a 5% CO2 atmosphere, using a live-cell instrument system (Chanlide® Inc., Korea). A confocal series of fluorescence and differential interference contrast (DIC) images were simultaneously obtained at 20-s intervals using an 100× oil immersion objective on FV1000 confocal microscope (Olympus). The images were processed and assembled into movies using Olympus FLUOVIEW software version 1.5.
Cell extract preparation and Western blot analysis
For the analysis of swiprosin-1, GFP, p-PI3K, and p-Akt HMC-1 cells (parent or transfected cells) were rinsed twice with ice-cold PBS and then lysed in ice-cold lysis buffer (10 mM Tris-HCl, pH 7.4, containing 50 mM NaCl, 1% Triton X-100, and a protease inhibitor cocktail tablet). Cell lysates were centrifuged at 14,000 rpm for 20 min at 4℃ and equal amounts of protein supernatant were mixed with a one-fourth volume of 4X SDS sample buffer, boiled for 5 min, and then separated through an 8 or 10% sodium dodecyl sulfate-polyacrylamide gel. After electrophoresis, proteins were transferred to a nitrocellulose membrane by means of the Trans-Blot SD semidry transfer cell (Bio-Rad, Hercules, CA). The membrane was blocked in 5% skim milk (1 h), rinsed, and incubated overnight at 4℃ with primary antibodies in TBS containing 0.1% Tween 20 (TBS-T) and 3% skim milk. Excess primary Ab was then removed by washing the membrane three times in TBS-T, and the membrane was incubated with 0.1µg/ml horseradish peroxidase-labeled secondary Ab (against goat, rabbit, or mouse) for 2 h. Following three washes in TBS-T, bands were visualized by ECL western blotting detection reagents and exposed to x-ray film.
IL-8 measurement and histamine release assay
Stable HMC-1 cells (H-GFP or H-swip-1_GFP; 2×106) were seeded into 12-well plates. The cells were further stimulated with PMA (200 nM)/A23187 (1µM). At the indicated time points, supernatants were collected and IL-8 and histamine levels were measured by ELISA (R&D Systems), according to the manufacturer's instructions.
Statistical analysis
The mean values were calculated from data taken from at least three (usually four or more) separate experiments conducted on separate days. Where significance testing was performed, an independent t test (Student's; two populations) was used. A p value of less than 0.05 was considered an indicator of statistical significance.
RESULTS
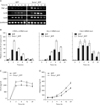
Swiprosin-1 is expressed in mast cells and up-regulated during mast cell activation
We examined the expression of swiprosin-1 in various cell types and found that it is expressed in most immune cells, including T and B cells, monocytes, and mast cells. It is also expressed in some non-immune cells, such as epithelial and endothelial cells, though there are some degrees of difference in terms of expression level (Fig. 1A). These results suggest that swiprosin-1 is not restricted to T and B cells but prevalently expressed in most cell types, including mast cells. With the semi-quantitative RT-PCR and real-time quantitative PCR, we confirmed that swiprosin-1 is significantly over-induced in HMC-1 cells when stimulated with PMA (100 nM)/A23187 (1 µM) (Fig. 1B). Swiprosin-1 was also over-induced in the rat mast cell line RBL-2H3 through the FcεRI cross-linking, by treatment with IgE and DNP-HSA, while the induction kinetics was faster than that of HMC-1 cells (2 to 6 h after stimulation) (Fig. 1C).
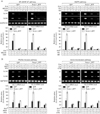
Swiprosin-1 enhances mast cell activation by phorbol ester and calcium ionophore
Induction of swiprosin-1 during mast cell activation, coupled with the fact that swiprosin-1 has a potential for a novel signaling adaptor protein (1,2,4,13), suggests that swiprosin-1 may be involved in mast cell activation as a signal adaptor molecule. To verify this, we generated HMC-1 cells that transiently or stably over-express either GFP (H-GFP cells) alone or GFP fused to swiprosin-1 (H-swip-1_GFP cells) (data not shown). As shown in Fig. 2A~C, over-expression of swiprosin-1 significantly enhanced PMA/A23187-induced cytokine expression. This result was consistent with the previous results (3). In addition, it enhanced histamine release by PMA/A23187 stimulation (Fig. 2D).
We next tested whether silencing swiprosin-1 can alter the cytokine expression in HMC-1 cells. To this end, we transfected HMC-1 cells with siRNA targeting swiprosin-1, and then evaluated the cells for cytokine expression. However, knockdown of swiprosin-1 only showed a modest effect on PMA/A23187-induced IL-3 and IL-8 mRNA expression in HMC-1 cells (data not shown), suggesting that swiprosin-1 has gain-of-function characteristics.
Cytochalasin B, an actin depolymerizing agent, and wortmannin, a PI3K inhibitor, selectively inhibit cytokine expression in swiprosin-1 expressing HMC-1 cells
The modest effect of swiprosin-1 knock-down on cytokine expression by PMA/A23187 may suggest that swiprosin-1 is not involved in the main pathway of cytokine expression but has an additive effect if over-expressed, presumably by modulating another pathway. We therefore employed various pharmacologic agents to systemically control the intracellular signaling pathways potentially involved in mast cell activation.
First, both NF-κB and NF-AT pathways are critical for IgE-mediated cytokine production (14,15). We examined the effects of two pharmacologic agents, MG132 and cyclosporine A, on PMA/A23187-induced cytokine expression in both H-GFP cells and H-swip-1_GFP cells. As shown in Fig. 3A (left), both of these agents significantly suppressed cytokine expression in both cells to almost the same degree. Second, MAPK signaling pathways are also involved in cytokine expression in mast cells (16-18). Thus, the effects of two pharmacologic inhibitors of ERK and p38 kinase, PD98059 (10µM) and SB203580 (10µM), respectively, were examined. As shown in Fig. 3A (right), both of the inhibitors significantly inhibited PMA/A23187-induced cytokine expression in both cells. Taken together, the above results suggest that swiprosin-1 lies at least upstream or at the site of two main signaling pathways in terms of cytokine expression in HMC-1 cells.
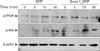
Third, both Src family kinases and PI3K are central players in mast cell degranulation mediated by the cross-linking of FcεR1 (7), but are not well understood in cytokine expression of HMC-1 cells mediated by phorbol ester and calcium ionophores. We therefore tested whether enhanced cytokine expression in H-swip-1_GFP is also related to these kinases. As shown in Fig. 3B (left), PP2, a Src kinase inhibitor, significantly inhibited cytokine expression in both H-GFP and H-swip-1_GFP cells, suggesting a requirement of Src family kinases in PMA/A23187-induced cytokine expression in HMC-1 cells. Interestingly, however, wortmannin, a PI3K inhibitor, only inhibited cytokine expression in Hswip-1_GFP cells (Fig. 3B, left). This result suggests that swiprosin-1 may influence cytokine expression in HMC-1 cells thorough activation of the PI3K pathway while the PI3K is not mainly involved in PMA/A23187-induced cytokine expression. Notably, we found that treatment with HMC-1 cells with PMA/A23187 resulted in no significant effect on the phosphorylation of PI3K, while p-Akt, a down-stream effector of PI3K, was slightly increased 5~30 min after treatment (Fig. 4). In contrast, both p-PI3K and p-Akt were observed in H-swip-1_GFP cells even before treatment (0 min) (Fig. 4).
Fourth, PI3K also has an important regulatory function for receptor-mediated cytoskeletal rearrangements in mast cells (19,20). We therefore evaluated the potential involvement of two of the most important components of the cytoskeleton, actin and microtubule. It has been reported that disruption of actin microfilaments by cytochalasin B enhanced the degranulation response in RBL-2H mast cells (21,22). In contrast, microtubule disruption is known to suppress the degranulation response through inhibition of calcium influx in the same cells (23). As shown in Fig. 3B (right), microtubule depolymerizer colchicine significantly suppressed cytokine expression in both H-GFP and H-swip-1_GFP cells, suggesting that microtubules are also important in PMA/A23187-induced cytokine expression in HMC-1 cells. However, actin depolymerizer cytochalasin B inhibited cytokine expression that was produced only in H-swip-1_GFP cells (Fig. 3B, right). These results demonstrate that actin polymerization is not normally involved in cytokine expression of HMC-1 cells stimulated by PMA/A23187. However, the present results strongly imply that swiprosin-1 may act through regulation of PI3K and actin regulation.
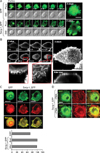
Swiprosin-1 localized in actin-rich microvilli-like region in mast cells
In many cases, localization of proteins in specialized sites within cells reflects their specific functions. Moreover, reduced cytokine expression in H-swip-1_GFP cells by actin depolymerizer cytochalasin B suggests a specific role of swiprosin-1 in association with the actin cytoskeleton (Fig. 3B, right). We therefore examined localization of swiprosin-1 in HMC-1 cells and other cell types using confocal microscopy. As shown in Fig. 5A, swip-1_GFP was highly localized in microvilli-like protrusions of HMC-1 cells, while GFP was distributed through the entire cytosol and nucleus. To better understand this contrast, wild-type swiprosin-1 (without GFP) was also transfected in 293T cells and stained with antibody against swiprosin-1 followed by FITC-conjugated secondary antibody. We then evaluated the localization by high resolution confocal microscopy. Interestingly, swiprosin-1 was notably accumulated in the membrane apical ridge area of 293T cells (Fig. 5B). This observation may agree with the report that swiprosin-1 is a lipid raft protein in B cells (4). Since F-actin is known to be enriched in the microvilli-like region (24), we tested whether swiprosin-1 is localized with F-actin in both HMC-1 and COS-7 cells. As shown in Fig. 5C and D, swiprosin-1 was highly co-localized with F-actin in both HMC-1 and COS-7 cells, and re-localized along with the movement of F-actin in COS-7 cells, when the cells were stimulated with PMA (Fig. 5D). Collectively, localization of swiprosin-1 in the microvilli-like region (Fig. 5A) or membrane ridge region (Fig. 5B) and its association with F-actin (Fig. 5C, D), together with the fact that the actin-disrupting agent cytochalasin B inhibits cytokine expression only in Hswip-1_GFP cells, suggest that swiprosin-1 functions through actin regulation and reorganization.
DISCUSSION
Molecules produced by mast cells also can activate mast cells in an autocrine manner. For example, TNF-α is one of the major cytokines that autocrinely activates HMC-1 cells (25). In a similar manner, ectopic expression of swiprosin-1 augmented PMA/A23187-induced NF-κB promoter activity as well as cytokine expression (3). In the present report, we demonstrate that localization of swiprosin-1 in microvilli-like membrane protrusion with F-actin may be important in activating mast cells because either disruption of actin by cytochalasin B or inhibition of PI3K blocked cytokine expression only in swiprosin-1-overexpressing cells.
In the previous study, we unambiguously demonstrated that swiprosin-1 expression is transcriptionally regulated both in vitro in a model of phorbol ester-induced activation and in vivo in pathological models. The results strongly suggested that swiprosin-1 may be importantly involved in physiological and/or pathological processes. Furthermore, increased expression of swiprosin-1 in tissue models of PCA and atopic dermatitis suggested a potential role for swiprosin-1 in inflammatory responses (3). With this respect, the current results strongly demonstrate that swiprosin-1 is involved in the inflammatory responses by expressing and releasing cytokines.
Localization of proteins in specialized sites in the cells can reflect their specific functions (26-28). For example, SLP-76, an adaptor protein that is critical for FcεRI-induced calcium flux, degranulation, and cytokine secretion in mast cells, is found in the cytosol of unstimulated cells. Upon FcεRI cross-linking, SLP-76 translocates to the cell membrane, forming clusters that co-localize with the FcεRI, the tyrosine kinase Syk, the adaptor LAT, and phosphotyrosine. Disruption of this localization has been reported to inhibit FcεRI-mediated signaling in mast cells (27). In accordance with this, the present finding that the highest concentration of swiprosin-1 in the microvilli-like membrane structure implies that this protein may function by forming signaling complexes in the plasma membrane or by associating with actin or actin regulatory molecules. Moreover, reduction of actin contents by over-expression of swiprosin-1 further supports its potential role in actin reorganization. Indeed, the proline-rich element in the predicted secondary structure of swiprosin-1 is known as a potential binding site for the SH3 domain (2), the best-characterized member of the growing family of protein-interaction modules involved in signaling to the dynamic actin networks (29,30). In accordance with the predicted secondary structure, our unpublished results revealed that deletion of the proline-rich element in swiprosin-1 significantly reduced its association with F-actin in COS-7 cells (data not shown).
Based on the evidence regarding the physical or functional association of swiprosin-1 with actin in the current study, it is not surprising that disruption of actin polymerization by cytochalasin B effectively inhibits cytokine expression in HMC-1 cells. Induction of p-PI3K and p-Akt and inhibition of cytokine expression by wortmannin also implies that swiprosin-1 affects the PI3K pathway coupled with the regulation of actin dynamics. However, the fact that targeted knockdown of swiprosin-1 showed a modest effect on cytokine expression may suggest that swiprosin-1 is not involved in the main pathways of cytokine expression but has an additive effect if over-induced, presumably activating actin reorganization in mast cells. We are now undertaking a study to identify the potential phosphorylation sites of swiprosin-1 and their implications in swiprosin-1-mediated activation of mast cells. In addition, we are planning to identify potential binding partners of swiprosin-1 to understand the mechanisms whether or how swiprosin-1 interacts with actin and regulates mast cell activation. One intriguing result in this study is that swiprosin-1 is rapidly (<5 min) and dramatically redistributed to the secreting granules of HMC-1 cells during activation by PMA/A23187 (data not shown). The result implies that swiprosin-1 may also be involved in the process of mast cell degranulation by coupling with actin dynamics.
The mast cell is a central player in allergy and asthma. Activation of these cells induces the release of preformed inflammatory mediators localized in specialized granules and the de novo synthesis and secretion of cytokines, chemokines, and eicosanoids (7). Upon stimulation, mast cell response can be regulated by the balance of both positive and negative intracellular molecular events (7). Here, we suggest that swiprosin-1 is a novel regulator for the positive feedback activation of mast cells. Once induced, this protein is highly localized in the actin-rich microvilli-like membrane protrusions in mast cells. In pathological situations, this protein may regulate local allergic or inflammatory responses by amplifying signals that are required for mast cell activation.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download