This article has been
cited by other articles in ScienceCentral.
Abstract
Objective
The aim of this study was to determine whether single nucleotide polymorphisms (SNPs) of fibroblast growth factor (FGF) 2 gene and fibroblast growth factor receptor (FGFR) genes are associated with ossification of the posterior longitudinal ligament (OPLL).
Methods
A total of 157 patients with OPLL and 222 controls were recruited for a case control association study investigating the relationship between SNPs of FGF2, FGFR1, FGFR2 and OPLL. To identify the association among polymorphisms of FGF2 gene, FGFR1, FGFR2 genes and OPLL, the authors genotyped 9 SNPs of the genes (FGF2 : rs1476217, rs308395, rs308397, and rs3747676; FGFR1 : rs13317 and rs2467531; FGFR2 : rs755793, rs1047100, and rs3135831) using direct sequencing method. SNPs data were analyzed using the SNPStats, SNPAnalyzer, Haploview, and Helixtree programs.
Results
Of the SNPs, a SNP (rs13317) in FGFR1 was significantly associated with the susceptibility of OPLL in the codominant (odds ratio=1.35, 95% confidence interval=1.01-1.81, p=0.048) and recessive model (odds ratio=2.00, 95% confidence interval=1.11-3.59, p=0.020). The analysis adjusted for associated condition showed that the SNP of rs1476217 (p=0.03), rs3747676 (p=0.01) polymorphisms in the FGF2 were associated with diffuse idiopathic skeletal hyperostosis (DISH) and rs1476217 (p=0.01) in the FGF2 was associated with ossification of the ligament flavum (OLF).
Conclusion
The results of the present study revealed that an FGFR1 SNP was significantly associated with OPLL and that a SNP in FGF2 was associated with conditions that were comorbid with OPLL (DISH and OLF).
Keywords: OPLL, FGF2 gene, FGFR 1 & 2 genes, Polymorphism
INTRODUCTION
Ossification of the posterior longitudinal ligament (OPLL) is an osteogenetic disorder of the spine that is common in East Asian populations. OPLL was first reported by Key
6) and later by Oppenheimer
13). However, this disease attracted attention as a cause of myelopathy only after the report by Tsukimoto in 1960
14,
15). The primary clinical feature of OPLL is ectopic bone formation in the posterior longitudinal ligament, which runs within the spinal canal. Patients develop compressive myelopathy and/or radiculopathy subsequently. The cause of this disease is unknown and the disorder is recognized as a common multifactorial disease
5,
14,
16). A variety of possible risk factors have been implicated in OPLL using epidemiological studies; these factors include sex, trauma, hormonal imbalance, dietary habits, and lifestyle factors (e.g., diabetes mellitus)
7). Genetic and environmental factors interact in OPLL and the genetic background is considered as a predominant factor in the etiology of OPLL
1,
5,
12,
16). Recent molecular genetic studies identified several candidate genes for susceptibility to this disease. These genes included collagen 11A2, BMP-4, nucleotide pyrophosphatase, TGF-β1, TGF-β3, ESR1, IL-1B, and retinoic X receptor; however, none of these genes have been confirmed as being pathogenetically relevant for OPLL patients
7,
16).
Bone formation throughout life is governed by osteoblasts, which are bone-forming cells. The rate of bone formation is dependent on the commitment and replication of osteoprogenitor cells, their differentiation into functional osteoblasts, and the life span of mature osteoblasts. Osteoblast differentiation is regulated by the actions of numerous systemic and local signaling factors. Among these factors, the important effects of fibroblast growth factors (FGFs) on the control of genes involved in the differentiation of osteoblasts have been recognized recently
3,
9). Thus, abnormalities of these FGF-related bone formation signaling pathways are closely related with ectopic ossification of spinal ligaments, such as OPLL, ossification of the ligament flavum (OLF), and diffuse idiopathic skeletal hyperostosis (DISH).
FGF-mediated bone formation is characterized by the replication of mesenchymal cells and differentiation of osteoprogenitor cells into mature osteoblasts and ends with osteoblast apoptosis. FGF acts via the fibroblast growth factor receptor (FGFR) to control several genes involved in osteoblast commitment, differentiation, and apoptosis
9).
Members of the FGF family of proteins play key roles in the growth and survival of stem cells during embryogenesis, tissue regeneration, and carcinogenesis. FGF signals are transduced via FGFRs, which include FGFR1, FGFR2, FGFR3, and FGFR4. These receptors contain an extracellular immunoglobulin-like domain and a cytoplasmic tyrosine kinase domain
2).
The FGF2 gene lies on chromosome 4 (at 4q25-q27). According to the Ensembl Gene View (Ensembl ID, Ensembl : ENSG 00000138685), it covers 71.5 kb from positions 123747863 to 123819390 on the forward strand. The FGF2 protein has been implicated in diverse biological processes, such as the development of limbs and nervous system, wound healing, and tumor growth. The FGFR1 gene lies on chromosome 8 (at 8p11.2p11.1). According to the Ensembl Gene View (Ensembl ID, Ensembl: ENSG00000077782), it covers 57.7 kb from positions 38268656 to 38326352 on the forward strand. This particular FGFR protein family member binds both acidic and basic FGFs and is involved in limb induction. The FGFR2 gene lies on chromosome 10 (at 10q26). According to the Ensembl Gene View (Ensembl ID, Ensembl : ENSG00000066468), it covers 12.0 kb from positions 123237848 to 123357972 on the forward strand. This FGFR protein family member is a high-affinity receptor for acidic, basic, and/or keratinocyte growth factors, depending on the isoform.
Recently, these FGF/FGFR signaling pathways have been found to play very important roles in bone development. Missense mutations in several FGF and FGFR genes were found in humans and cause a variety of congenital bone diseases, including craniosynostosis syndromes, chondrodysplasia syndromes, and syndromes with dysregulated phosphate metabolism
3,
9,
10,
11). In addition to its role in bone development and in genetic diseases, FGF signaling is also involved in the maintenance of adult bone homeostasis, fracture healing, primary ligament repair, etc.
The fact that FGFs/FGFRs play a major role in bone development led us to suspect that it may also play an important role in the pathogenesis of human OPLL. Nevertheless, there are no reports on the genetic association of these genes with the pathogenesis of OPLL.
The purpose of this study was to investigate the possible relationship between polymorphisms in the FGF2, FGFR1, and FGFR2 genes and OPLL in Korean populations.
MATERIALS AND METHODS
Study Population
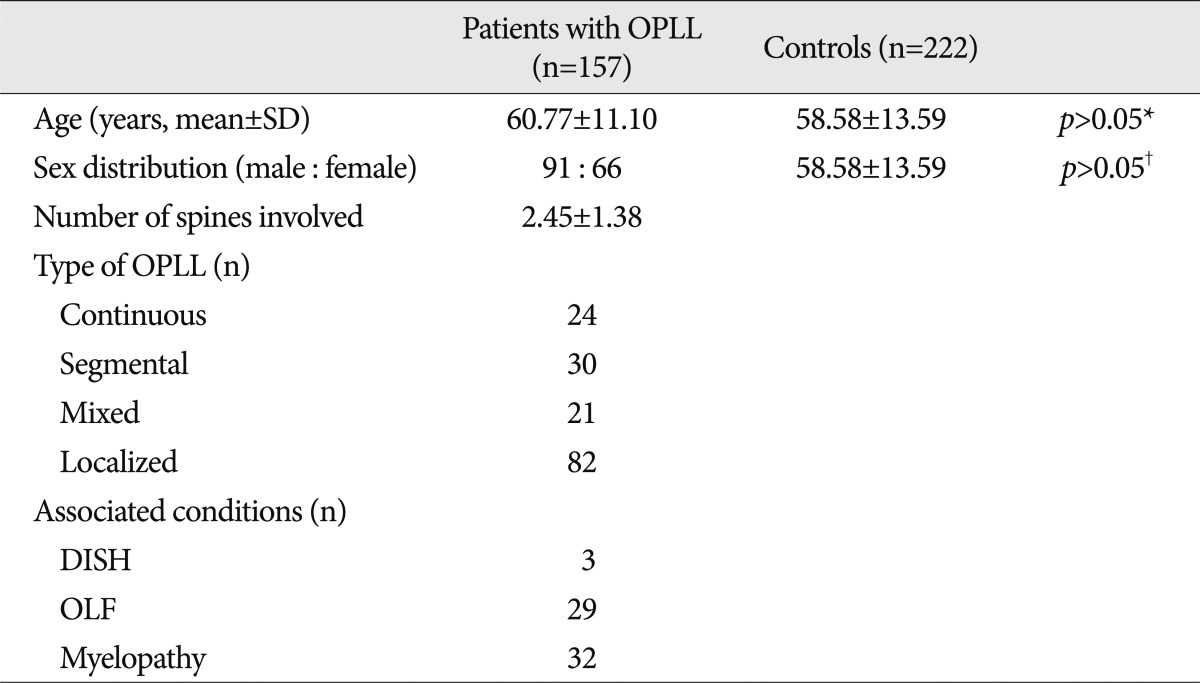
The OPLL patients were diagnosed using computed tomography, magnetic resonance imaging, and/or radiographic findings. The OPLL group included 157 patients (91 males and 66 females; mean age, 60.8±11.1 years). Two-hundred-twenty-two healthy control individuals (110 males and 112 females; mean age, 58.6±13.6 years) were recruited after confirmation via a general health check-up program that they had no clinical and radiological (imaging studies were done by simple radiographs and computed tomography) evidence of OPLL or any other disorders (
Table 1).
Informed consent was obtained from all individuals, according to the Declaration of Helsinki guidelines. This study protocol was approved by the Ethics Committee of the Medical Research Institute, School of Medicine, Kyung Hee University, Seoul, Korea. Blood samples for DNA extraction were collected from all subjects using ethylenediamine tetraacetic acid tubes. Genomic DNA was extracted from 100 µL of whole blood using a DNA isolation kit (NucleoSpin®) for mammalian blood.
Single-nucleotide polymorphism selection and genotyping
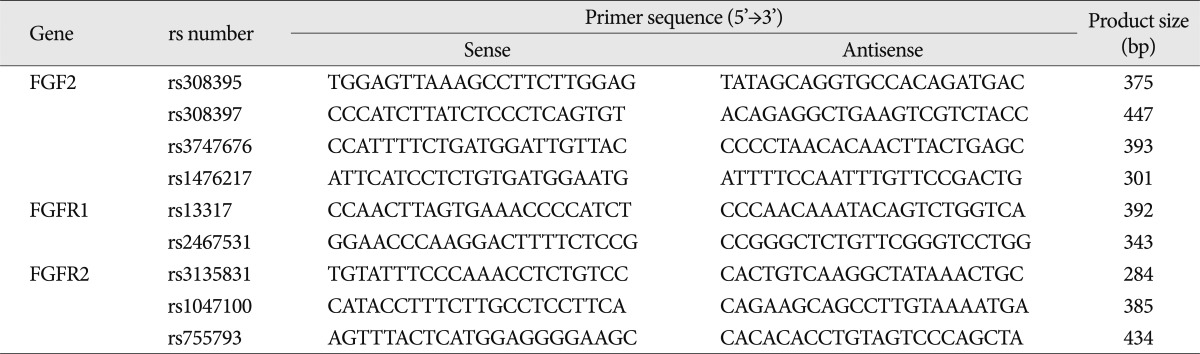
Four single-nucleotide polymorphisms (SNPs) within the FGF2 gene, two SNPs within the FGFR1 gene, and three SNPs within the FGFR2 gene were selected using online human SNP databases (
http://www.ensembl.org;
http://www.ncbi.nlm.nih.gov/SNP). SNPs with unknown heterozygosity and minor allele frequencies below 5% were excluded.
Genomic DNA was amplified using primers that were specific for each SNP (
Table 2). PCR products were sequenced using the ABI Prism Big Dye Terminator Cycle Sequencing system (PE Applied Biosystems, Foster City, CA, USA) and visualized on 6% polyacrylamide/6M urea sequencing gels using an ABI Prism 377 automatic sequencer (PE Applied Biosystems, Foster City, CA, USA). The sequence data were analyzed using the SeqManII software (DNASTAR Inc., Madison, WI, USA).
Statistical analyses
Statistical analyses were performed using the SPSS software (version 17.0, Chicago, IL, USA).
The chi-squared (χ
2) test was used to evaluate Hardy-Weinberg equilibrium (HWE) between each genotype and each individual. Single SNP analyses were performed using the SNPAnalyzer and SNPStats software (
http://bioinfo.iconcologia.net/index.php). SNP effects were analyzed using codominant, dominant, and recessive models. Logistic regression models were calculated for odds ratios (OR), 95% confidence intervals (CIs), and corresponding
p values, while controlling for gender as a covariable. Significance was set at
p<0.05 for all statistical tests.
To evaluate the presence of linkage disequilibrium (LD) and blocks of haplotypes between polymorphisms within the FGF2, FGFR1, and FGFR2 genes, LD and haplotypes were analyzed using the Haploview software, version 4.1.
RESULTS
In this study, we investigated whether polymorphisms within FGF2, FGFR1, and FGFR2 were associated with OPLL by genotyping the nine selected SNPs in Korean populations.
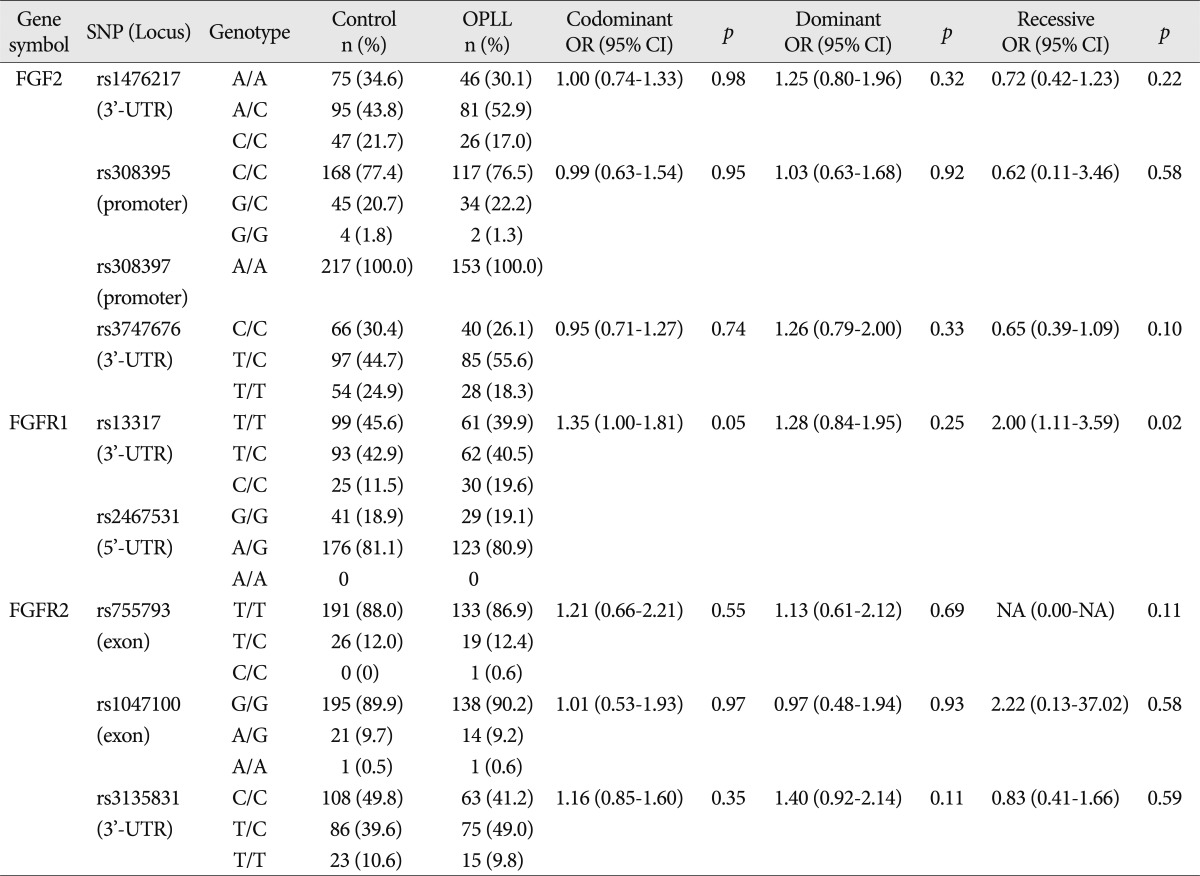
Among the nine SNPs examined, seven were polymorphic (rs1476217, rs308395, rs3747676, rs13317, rs755793, rs1047100, and rs3135831). The genotype distribution of these seven SNPs was in HWE (p>0.05). The four SNPs selected among the various SNPs present in the FGF2 gene included rs1476217 (3'-UTR), rs308395 (promoter), rs308397 (promoter), and rs3747676 (3'-UTR). Among these SNPs, rs308397 was excluded from the analysis because it showed specific A/A monomorphism (not in HWE; p<0.05). The genotype frequency of the three SNPs of FGF2 was compared between the OPLL patients and the controls using logistic regression models. Logistic regression analysis controlling for age and sex as covariables in all three analytical models (codominant, dominant, and recessive models for rare alleles) was used to assess alternative effects of the variants; however there were no significant differences between OPLL and controls regarding the three SNPs of FGF2. The two FGFR1 SNPs included in the analysis were rs13317 (3'-UTR) and rs2467531 (5'-UTR). rs2467531 was excluded from the analysis because the frequency of the A/A genotype was 0%. Logistic regression analysis controlling for age and sex as covariables in all three analytical models (codominant, dominant, and recessive models for rare alleles) was also employed and revealed an absence of significant differences between the OPLL and control groups regarding the FGFR1 SNP (rs13317, p=0.048, OR=1.35, 95% CI=1.00-1.81 for the codominant model; p=0.020, OR=2.00, 95% CI=1.11-3.59 for the recessive model).
The three FGFR2 SNPs included in the analysis were rs755793 (exonic), rs1047100 (exonic), and rs3135831 (3'-UTR). No deviation of HWE was found in the genotype distributions of FGFR2 SNPs. All these SNPs were analyzed using the statistical method described above; there were no significant differences between OPLL and control subjects (
Table 3).
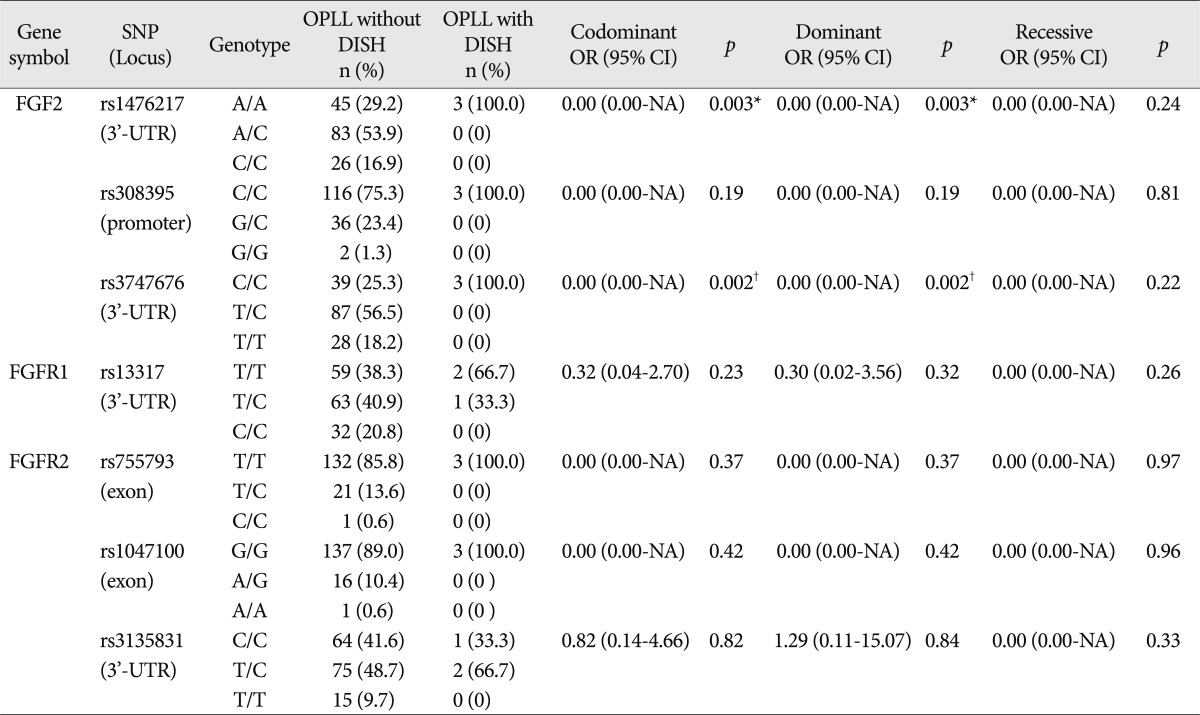
We also analyzed whether the conditions associated with OPLL (myelopathy, DISH, and OLF) were associated with FGF2, FGFR1, and FGFR2 polymorphisms. The comparison of the distributions of genotypes between OPLL patients groups presenting with and without myelopathy revealed an absence of significant differences between the groups. The distribution of genotypes between OPLL patients groups presenting with and longiwithout DISH revealed that two SNPs of FGF2 exhibited a significant association [rs1476217,
p=0.03 (Fisher exact test); rs3747676,
p=0.01 (Fisher exact test)] (
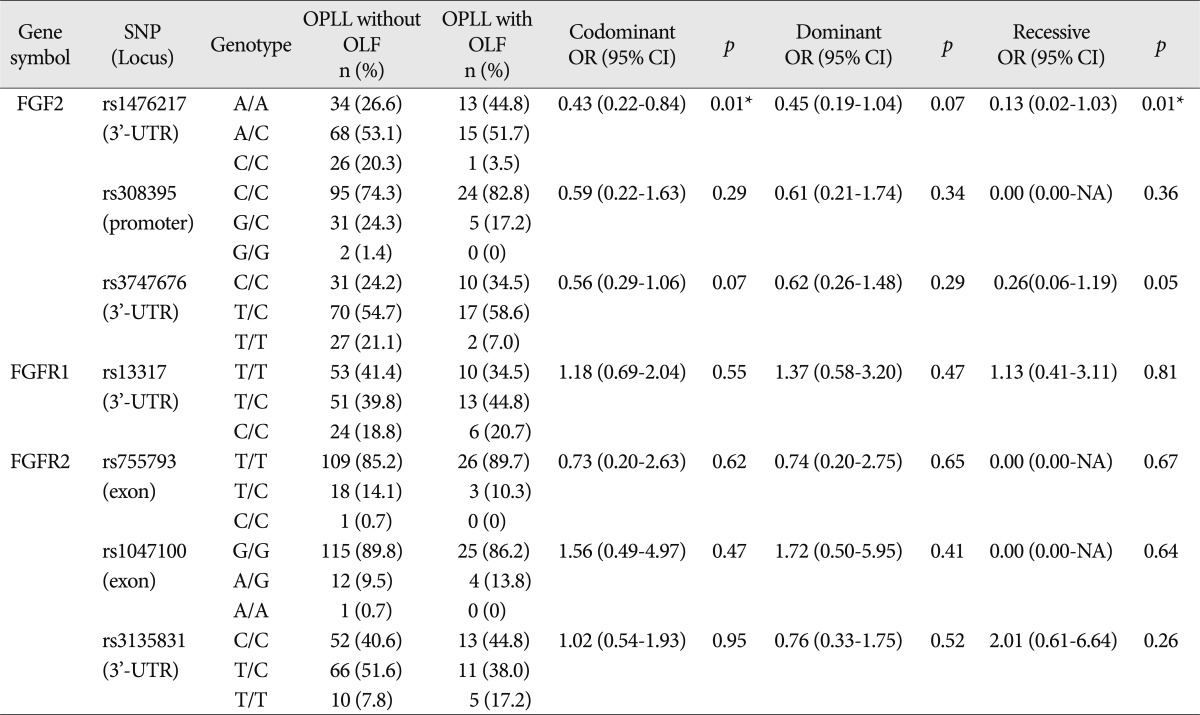
Table 4). Genotype frequency analysis between OPLL patients groups presenting with and without OLF showed that one SNP of FGF2 exhibited a significant difference between the groups [rs1476217,
p=0.01 (Fisher exact test)] (
Table 5).
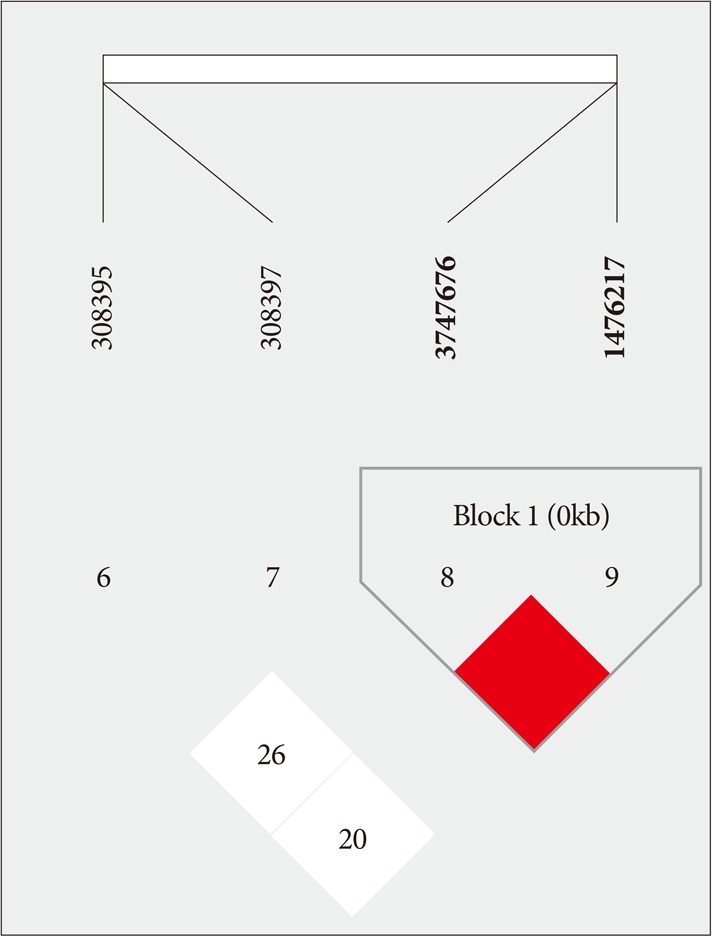
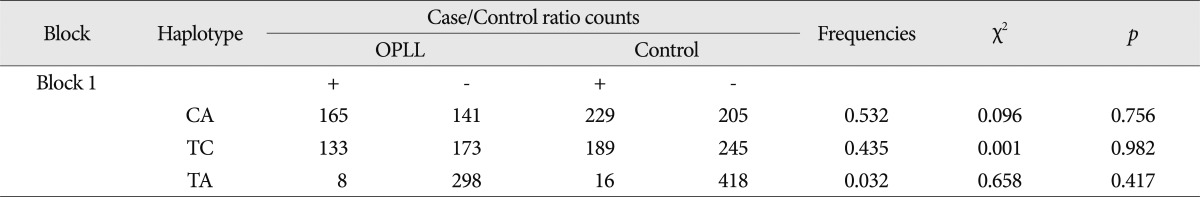
According to the LD and haplotype analyses, one LD block was observed for four SNPs within the FGF2 gene, as assessed using the Gabriel method. Haplotypes were constructed in block 1 (
Fig. 1). Haplotypes of block 1 included rs3747676 and rs1476217; however, there was no significant difference between OPLL subjects and controls regarding this block (
Table 6).
DISCUSSION
Many studies have been performed to determine the cause of OPLL which led to the identification of general and local factors that seemed correlated with disease etiology. As OPLL is frequently complicated by ossification of the anterior longitudinal and yellow ligament, some researchers assumed that OPLL is a phenotype of osteosis of the spinal ligaments
8). Abnormal metabolism of hormones and calcium, diabetes, and degeneration are also said to be involved in this disease. In addition, many local factors, such as mechanical stress and trauma, have been proposed. Among the recent reports on the cause of OPLL, examination of the genetic background of the patients has been attracting attention. The existence of genetic factors that underlie the pathogenesis of OPLL has been suggested by the results of pedigree (OPLL was found in 30% of siblings of OPLL patients) and twin (OPLL was found in the siblings of six among eight identical twins) studies performed in Japan
4,
12). The association of OPLL susceptibility with particular human leukocyte antigen haplotypes on chromosome 6, which was reported by Sakou et al.
14), led to the hypothesis that genetic factors located in, or associated with, chromosome 6 may be involved in the etiology of the disease. The detection of specific polymorphisms located in genes that regulate the metabolism of bone, cartilage, or ligaments found at a significantly higher (or lower) frequency in patients with OPLL compared with control individuals may imply that these polymorphisms have a causal association with OPLL.
According to Sakou et al.
14), abnormalities of physical properties or physiological functions may occur in the posterior longitudinal ligament of OPLL patients, which may alter the homeostasis of the ligament tissue and lead to the loss of normal repair functions. Eventually, inflammation may occur and abnormal repair and ossification of ligament tissue will take place because of a genetic predisposition.
Numerous studies have implicated FGFs as key molecules during the initiation of the cellular proliferation, differentiation, migration, and matrix deposition that characterize wound healing
3,
9,
11). FGFs are also known as hemostatic factors that function in tissue repair and in response to injury. In addition, they have been implicated as important regulators of bone metabolism and are present in the ossified matrix and chondrocytes of cartilage that is adjacent to areas of OPLL. Therefore, FGFs may play an important role in the development of OPLL.
The human FGF gene family comprises at least 23 different genes that encode related polypeptides. FGFs are expressed in almost all tissues and play important roles in a variety of normal and pathological processes, including development, wound healing, and neoplastic transformation
9). FGFs interact with a family of four distinct, high-affinity tyrosine kinase receptors, the FGFRs. The FGFR genes constitute a gene family of four membrane-bound receptor tyrosine kinases (FGFR1-4) that mediate the signals of at least 22 FGFs (FGF1-22). FGFs/FGFRs play important roles in multiple biological processes, including mesoderm induction and patterning, cell growth and migration, organ formation, and bone growth. Furthermore, it has been shown that missense mutations in FGFR1-3 in humans result in congenital bone diseases
2).
In this study, we investigated the association between FGF2, FGFR1, and FGFR2 polymorphisms and OPLL. We found that FGFR1 was associated with OPLL in Korean patients. In a case-control analysis, one SNP (rs13317) examined in FGFR1 was significantly associated with OPLL. This means that the population that carried the CC genotype had a probability of developing OPLL that was twice that observed for controls. However, we did not find an association between FGF2 and FGFR2 and OPLL. Analysis of the data adjusting for associated conditions showed that the rs1476217 and rs3747676 polymorphisms in the FGF2 gene were associated with DISH and that the rs 1476217 polymorphism in the FGF2 gene was associated with OLF.
The human SNP database (
www.ncbi.nlm.nih.gov/SNP, dbSNP Build 130) includes the genotype frequencies for rs1476217 (A/A : A/C : C/C, European : 0.283 : 0.550 : 0.167; Chinese : 0.444 : 0.489 : 0.067; Japanese : 0.273 : 0.477 : 0.250; Sub-Saharan African : 0.217 : 0.417 : 0.367), rs308395 (C/C : C/G : G/G, European : 0.767 : 0.183 : 0.050; Chinese : 0.889 : 0.111 : 0.000; Japanese : 0.733 : 0.267 : 0.000; Sub-Saharan African : 0.083 : 0.350 : 0.567), rs308397 (Global, A allele : 0.941, G allele : 0.059), rs3747676 (C/C : C/T : T/T, European : 0.200 : 0.567 : 0.233; Chinese : 0.289 : 0.622 : 0.089; Japanese : 0.227 : 0.432 : 0.341; Sub-Saharan African : 0.000 : 0.033 : 0.967), rs13317 (C/C : C/T : T/T, European : 0.067 : 0.383 : 0.550; Chinese : 0.111 : 0.511 : 0.378; Japanese : 0.136 : 0.432 : 0.432; Sub-Saharan African : 0.017 : 0.200 : 0.783), rs2467531 (A/A : A/G : G/G, Global : 0.000 : 0.090 : 0.910), rs755793 (C/C : C/T : T/T, European : 0.000 : 1.000 : 0.000; Chinese : 0.000 : 1.000 : 0.000; Japanese : 0.000 : 1.000 : 0.000; Sub-Saharan African : 0.000 : 1.000 : 0.000), rs1047100 (A/A : A/G : G/G, European : 0.067 : 0.350 : 0.583; Chinese: 0.000 : 0.111 : 0.889; Japanese : 0.023:0.159 : 0.818; Sub-Saharan African : 0.050 : 0.417 : 0.533), and rs3135831 (C/C : C/T : T/T, European : 0.283 : 0.600 : 0.117; Chinese : 0.422 : 0.511 : 0.067; Japanese : 0.600 : 0.333 : 0.067; Sub-Saharan African : 0.983 : 0.017 : 0.000). The genotype distributions of all the SNPs analyzed in the control group were similar to those of Asian populations, and especially to those of the Japanese population.
CONCLUSION
In conclusion, the results of the present study revealed that an FGFR1 SNP was significantly associated with OPLL and that a SNP in FGF2 was associated with conditions that were comorbid with OPLL (DISH and OLF). To our knowledge, this is the first report on the assessment of the association of polymorphism of FGF and FGFR genes and OPLL. As no other association studies regarding FGF and FGFR genes and OPLL are available at the present time, additional studies are required to confirm the possible role of SNPs located in FGF and FGFR genes in the development of OPLL.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download