Abstract
Purpose:
Mounting evidence indicates that perturbation of tyrosine phos-phorylation is implicated in the development of many human diseases, including cancers. Docking proteins (DOKs) are tyrosine-phosphorylated proteins that negatively regulate tyrosine kinase signaling and they are considered to be tumor suppressors. Deletion and the altered expression of the DOK2 gene have been studied in leukemias and lung cancers. However, the somatic mutation status of the DOK2 gene has not been studied in lung cancers. The aim of this study was to see whether alterations of DOK2 protein expression and somatic mutation of the DOK2 gene are present in human non-small cell lung cancer (NSCLC). Materials and Methods: We analyzed DOK2 somatic mutation in 45 NSCLCs (23 adenocarcinomas (AD) and 22 squamous cell carcinomas (SCC) by single-strand conformation polymorphism (SSCP). We examined the DOK2 protein expression in 45 NSCLCs by immunohistochemistry.
Results:
SSCP analysis revealed no evidence of somatic mutation in the DNA sequences encoding the DOK2 gene in the 45 NSCLCs. Among the informative cases, 27% and 21% of the ADs and SCCs showed allelic loss in the DOK2 locus, respectively. On the immunohistochemistry, DOK2 protein was expressed in the normal bronchial epithelial cells, while it was lost in 10 (22%) of the NSCLCs.
Conclusion: Our data indicates that DOK2 is altered in NSCLC at the expressional level, but not at the mutational level. The data also suggests that loss of the expression of DOK2 might play roles in NSCLC development by possibly altering tyrosine kinase signaling.
REFERENCES
1. Gschwind A, Fischer OM, Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat Rev Cancer. 2004; 4:361–370.

2. Neel BG, Tonks NK. Protein tyrosine phosphatases in signal transduction. Curr Opin Cell Biol. 1997; 9:193–204.

3. Krause DS, Van Etten RA. Tyrosine kinases as targets for cancer therapy. N Engl J Med. 2005; 353:172–187.

5. Di Cristofano A, Carpino N, Dunant N, et al. Molecular cloning and characterization of p56dok-2 defines a new family of RasGAP-binding proteins. J Biol Chem. 1998; 273:4827–4830.
6. Suzu S, Tanaka-Douzono M, Nomaguchi K, et al. p56(dok-2) as a cytokine-inducible inhibitor of cell proliferation and signal transduction. EMBO J. 2000; 19:5114–5122.

7. Grimm J, Sachs M, Britsch S, et al. Novel p62dok family members, dok-4 and dok-5, are substrates of the c-Ret receptor tyrosine kinase and mediate neuronal differentiation. J Cell Biol. 2001; 154:345–354.

8. Dong S, Corre B, Foulon E, et al. T cell receptor for antigen induces linker for activation of T cell-dependent activation of a negative signaling complex involving Dok-2, SHIP-1, and Grb-2. J Exp Med. 2006; 203:2509–2518.

9. Gé rard A, Favre C, Garç on F, et al. Functional interaction of RasGAP-binding proteins Dok-1 and Dok-2 with the Tec protein tyrosine kinase. Oncogene. 2004; 23:1594–1598.

10. Cong F, Yuan B, Goff SP. Characterization of a novel member of the DOK family that binds and modulates Abl signaling. Mol Cell Biol. 1999; 19:8314–8325.

11. Jones N, Dumont DJ. Recruitment of Dok-R to the EGF receptor through its PTB domain is required for attenuation of Erk MAP kinase activation. Curr Biol. 1999; 9:1057–1060.

12. Niki M, Di Cristofano A, Zhao M, et al. Role of Dok-1 and Dok-2 in leukemia suppression. J Exp Med. 2004; 200:1689–1695.

13. Mashima R, Hishida Y, Tezuka T, Yamanashi Y. The roles of Dok family adapters in immunoreceptor signaling. Immunol Rev. 2009; 232:273–285.

14. Yasuda T, Shirakata M, Iwama A, et al. Role of Dok-1 and Dok-2 in myeloid homeostasis and suppression of leukemia. J Exp Med. 2004; 200:1681–1687.

15. Berger AH, Niki M, Morotti A, et al. Identification of DOK genes as lung tumor suppressors. Nat Genet. 2010; 42:216–223.

16. Berger AH, Pandolfi PP. Haplo-insufficiency: a driving force in cancer. J Pathol. 2011; 223:137–146.

17. Lee SH, Shin MS, Park WS, et al. Alterations of Fas (Apo-1/CD95) gene in non-small cell lung cancer. Oncogene. 1999; 18:3754–3760.

18. Lee JW, Soung YH, Kim SY, et al. PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene. 2005; 24:1477–1480.

Fig. 1.
Representative SSCP of DOK2 gene in lung cancers. Exon 5 of DOK2 gene was amplified by polymerase chain reaction (PCR) using a specific primer set. The PCR products from 5 representative cases of non-small cell lung cancers were visualized on SSCP. SSCP of DNA from the non-small cell lung cancers (T) shows no aberrant bands as compared to SSCPs from the normal tissues (N). DOK: docking proteins, SSCP: single-strand conformation polymorphism.
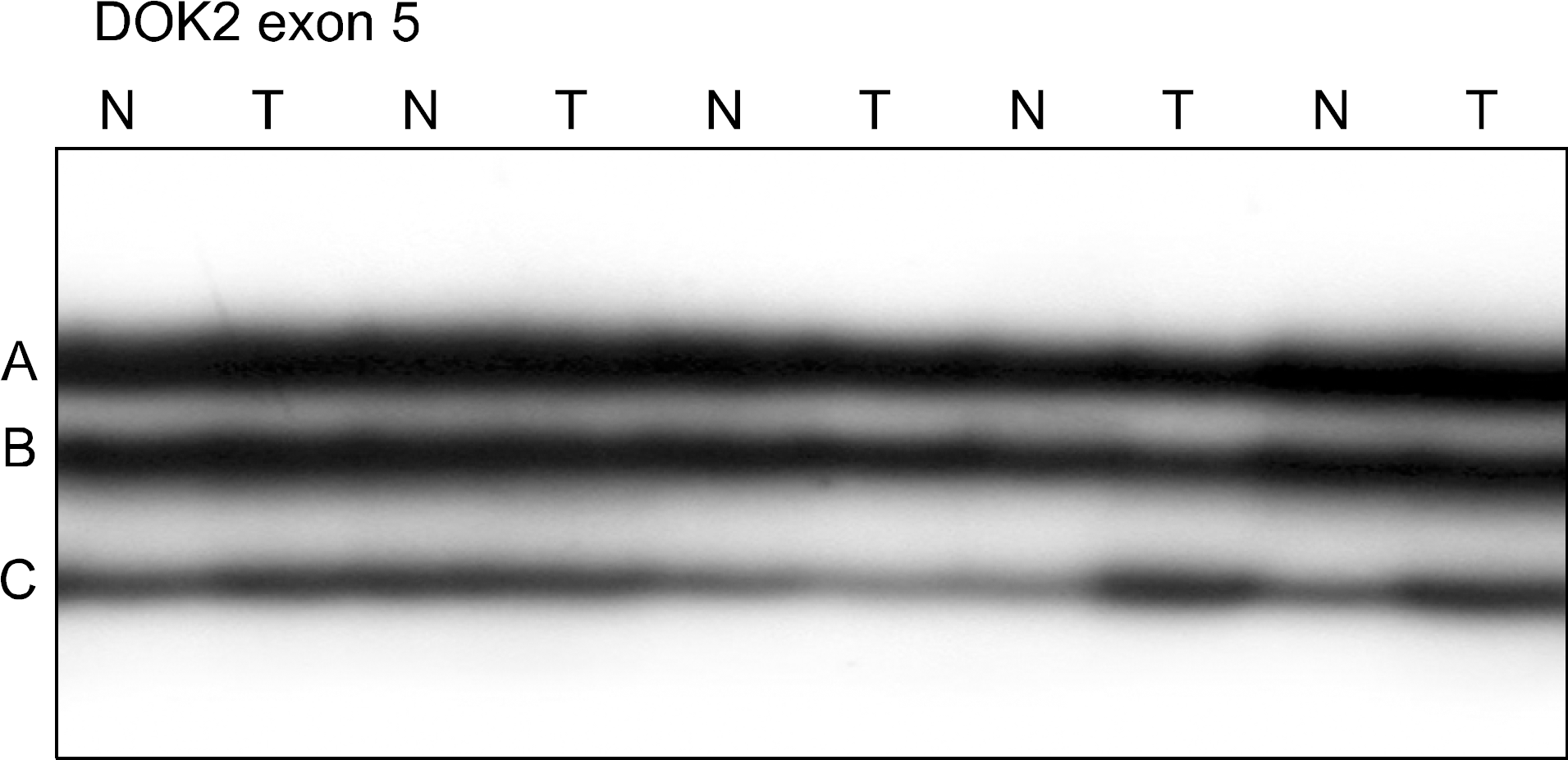
Fig. 2.
Visualization of DOK2 expression in lung cancer tissues by immunohistochemistry. (A) A nest of a squamous cell carcinoma (T) shows DOK2 immunostaining in the cancer cells. (B) Glands of an adenocarcinoma shows DOK2 immunostaining in the cancer cells (T). (C) In another adenocarcinoma, the cancer cells in the glands (T) are negative for DOK2 immunostaining. (D) Normal bronchial epithelial cells (arrow) are positive for DOK2 immunostaining (original magnification A, B, D, ×200; C, ×100).
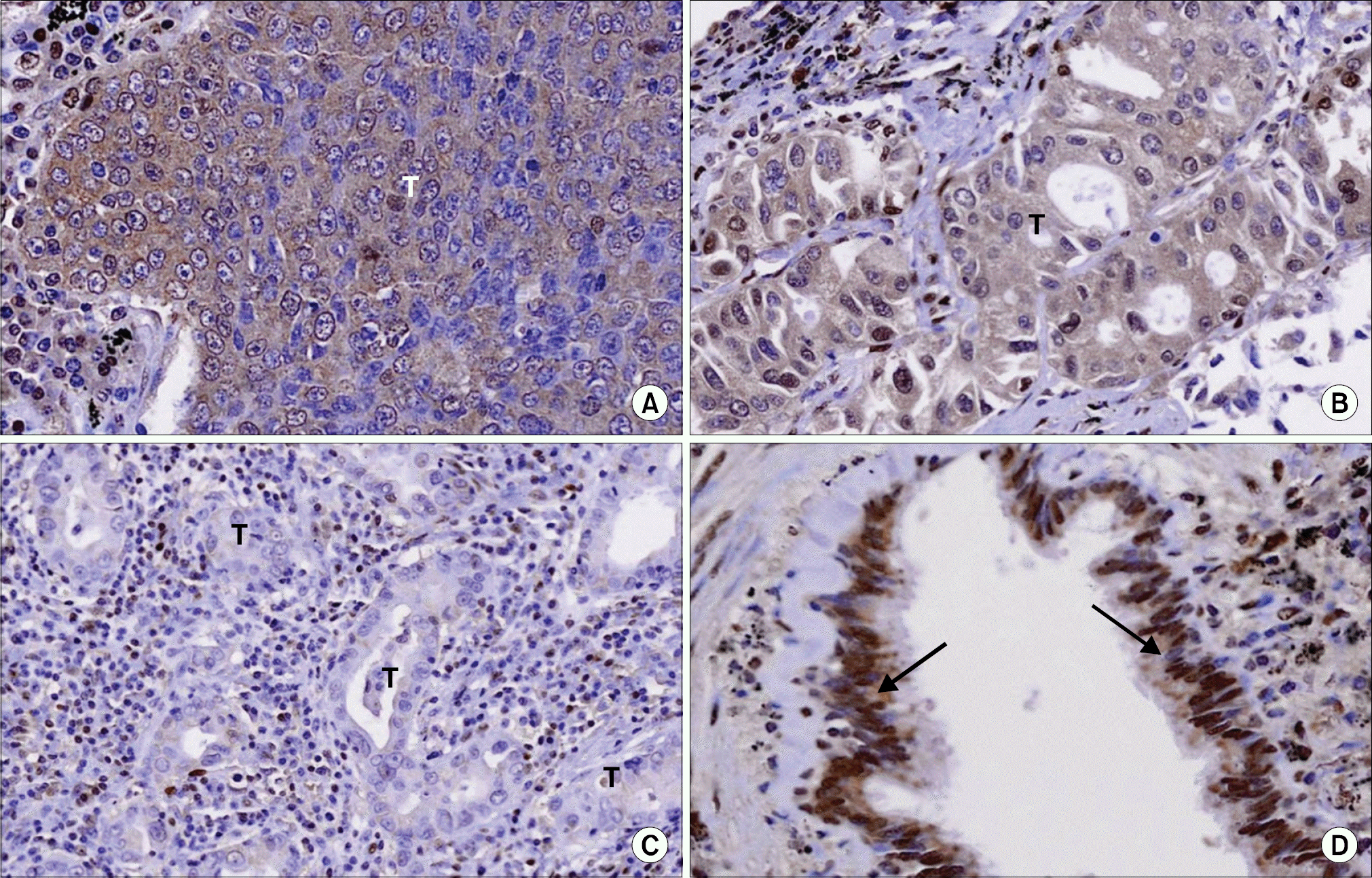
Table 1.
Primer Sequences of DOK2 Gene Used in the SSCP
Table 2.
LOH of DOK2 Gene in Lung Cancers Identified by Microsatellite Markers and Intragenic Polymorphisms
| Gene | Markers | Distance away from the gene | No. of AD with LOH∗ | No. of SCC with LOH∗ |
|---|---|---|---|---|
| DOK2 | rs2242240 | Intragenic (exon 5) | 4 (27) | 3 (21) |
| rs17853066 | Intragenic (exon 5) | |||
| rs899428 | Intragenic (intron 1) | |||
| AFM234YH10 | 4 kb | |||
| AFMA127YE5 | 164 kb |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download