Abstract
Background
We evaluated multiplex PCR for species identification and toxin typing to improve the sensitivity and turnaround time of toxigenic Clostridium difficile culture (TCDC).
Methods
We performed multiplex PCR using primers targeting the species-specific gene, tpi, and the toxin genes, tcdA and tcdB. From January to March 2008, 528 stool specimens were tested with direct toxin assay (DT) using C. difficile Tox A/B II (Techlab, Blacksburg, USA) and TCDC. For 288 specimens from early study period, toxin production by C. difficile isolates of TCDC was measured by enzyme immunoassay with culture supernatants using VIDAS C. difficile Toxin A&B (CDAB; bioMérieux, Marcy-l'Etoile, France) and multiplex PCR with isolated colonies. For 240 specimens from late period, only multiplex PCR was used to test toxin production by the isolates.
Results
During the early period, 29 C. difficile were isolated and their toxin-positive rates were 65.5% by PCR and 44.8% by CDAB (P<0.05). Among 528 stool specimens, the results of DT+/TCDC+, DT+/ TCDC-, and DT-/TCDC+ were 32 (6.1%), 33 (6.3%), and 10 (1.9%), respectively, when tested with PCR. 13.3% of total 75 positive specimens was detected only by TCDC. Of the 42 toxigenic C. difficile isolates, all were positive for tpi, 30 (71.4%) were tcdA+/tcdB+, and 12 (28.6%) were tcdA-/tcdB+.
REFERENCES
1. Moncrief JS, Zheng L, Neville LM, Lyerly DM. Genetic characterization of toxin A-negative, toxin B-positive Clostridium difficile isolates by PCR. J Clin Microbiol. 2000; 38:3072–5.
2. Delmée M, Van Broeck J, Simon A, Janssens M, Avesani V. Laboratory diagnosis of Clostridium difficile-associated diarrhoea: a plea for culture. J Med Microbiol. 2005; 54:187–91.
3. Reller ME, Lema CA, Perl TM, Cai M, Ross TL, Speck KA, et al. Yield of stool culture with isolate toxin testing versus a two-step algorithm including stool toxin testing for detection of toxigenic Clostridium difficile. J Clin Microbiol. 2007; 45:3601–5.
4. Park HK, Lee YM, Jang HJ, Kim CM, Lee K, Jeong SH, et al. Direct detection of Clostridium difficile toxin B gene by nested PCR in human stool specimens. Korean J Clin Microbiol. 2003; 6:63–8.
5. Fedorko DP, Williams EC. Use of cycloserine-cefoxitin-fructose agar and L-proline-aminopeptidase (PRO Discs) in the rapid identification of Clostridium difficile. J Clin Microbiol. 1997; 35:1258–9.
6. Kim H, Riley TV, Kim M, Kim CK, Yong D, Lee K, et al. Increasing prevalence of toxin A-negative, toxin B-positive isolates of Clostridium difficile in Korea: impact on laboratory diagnosis. J Clin Microbiol. 2008; 46:1116–7.
7. Shin BM, Kuak EY, Yoo SJ, Shin WC, Yoo HM. Emerging toxin A-B+ variant strain of Clostridium difficile responsible for pseudomembranous colitis at a tertiary care hospital in Korea. Diagn Microbiol Infect Dis. 2008; 60:333–7.
8. Shin BM, Kuak EY. Characterization of a toxin A-negative, toxin B-positive variant strain of Clostridium difficile. Korean J Lab Med. 2006; 26:27–31.
9. Lemee L, Dhalluin A, Testelin S, Mattrat MA, Maillard K, Lemeland JF, et al. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (Toxin A), and tcdB (Toxin B) genes for toxigenic culture of Clostridium difficile. J Clin Microbiol. 2004; 42:5710–4.
10. bioMérieux. VIDAS C. difficile Toxin A&B manual. Marcy-l'Etoile, France,. 2007; 7.
11. Chouicha N, Marks SL. Evaluation of five enzyme immunoassays compared with the cytotoxicity assay for diagnosis of Clostridium difficile-associated diarrhea in dogs. J Vet Diagn Invest. 2006; 18:182–8.
12. Kang JO, Chae JD, Eom JI, Han DS, Park PW, Park IK, et al. Comparison of Clostridium difficile toxin A immunoassay with cytotoxicity assay. Korean J Clin Microbiol. 2000; 3:43–7.
13. Murray P. ed. Manual of Clinical Microbiology. 9th ed. Washington; ASM Press, 2007: 899.
14. Kamiya S, Yamakawa K, Ogura H, Nakamura S. Effect of various sodium taurocholate preparations on the recovery of Clostridium difficile spores. Microbiol Immunol. 1987; 31:1117–20.
15. BD Diagnostic Systems ed. Difco & BBL Manual. 1st ed. Sparks, MD USA, 2003: 148.
16. Garcia A, Garcia T, Pérez JL. Proline-aminopeptidase test for rapid screening of Clostridium difficile. J Clin Microbiol. 1997; 35:3007.
17. Elsayed S, Zhang K. Positive Clostridium difficile stool assay in a patient with fatal C. sordellii infection. N Engl J Med. 2006; 355:1284–5.
18. Blake JE, Mitsikosta F, Metcalfe MA. Immunological detection and cytotoxic properties of toxins from toxin A-positive, toxin B-positive Clostridium difficile variants. J Med Microbiol. 2004; 53:197–205.
19. van den Berg RJ, Claas EC, Oyib DH, Klaassen CH, Dijkshoorn L, Brazier JS, et al. Characterization of toxin A-negative, toxin B-positive Clostridium difficile isolates from outbreaks in different countries by amplified fragment length polymorphism and PCR ribotyping. J Clin Microbiol. 2004; 42:1035–41.
20. Ricciardi R, Rothenberger DA, Madoff RD, Baxter NN. Increasing prevalence and severity of Clostridium difficile colitis in hospitalized patients in the United States. Arch Surg. 2007; 142:624–31.
21. Lyerly DM, Neville LM, Evans DT, Fill J, Allen S, Greene W, et al. Multicenter evaluation of the Clostridium difficile TOX A/B TEST. J Clin Microbiol. 1998; 36:184–90.
22. Brazier JS, Stubbs SL, Duerden BI. Prevalence of toxin A negative/B positive Clostridium difficile strains. J Hosp Infect. 1999; 42:248–9.
23. Barbut F, Lalande V, Burghoffer B, Thien HV, Grimprel E, Petit JC. Prevalence and genetic characterization of toxin A variant strains of Clostridium difficile among adults and children with diarrhea in France. J Clin Microbiol. 2002; 40:2079–83.
24. Komatsu M, Kato H, Aihara M, Shimakawa K, Iwasaki M, Nagasaka Y, et al. High frequency of antibiotic-associated diarrhea due to toxin A-negative, toxin B-positive Clostridium difficile in a hospital in Japan and risk factors for infection. Eur J Clin Microbiol Infect Dis. 2003; 22:525–9.
25. Legaria MC, Lumelsky G, Rosetti S. Clostridium difficile-associated diarrhea from a general hospital in Argentina. Anaerobe. 2003; 9:113–6.
Fig. 1.
Electrophoresis of the PCR products of nine isolates of C. difficile using DNA templates extracted with GenElute bacterial genomic DNA kit (Sigma-Aldrich, St. Louis, USA) (A) and boiling method (B) in parallel. The bands on all lanes of A are consistently denser than those of B except lane 3. Lane M; 100 bp DNA ladder, Lane 1; negative control, Lane 2 and 3; positive for species specific gene (tpi) but negative for toxin genes (tcdA and tcdB), indicating nontoxigenic strains, Lane 4-7; positive for tpi, tcdA, and tcdB genes, indicating A+B+ toxigenic strains, Land 8-10; positive for tpi, and tcdB, but negative (deleted) for tcdA gene, indicating A-B+ toxigenic strains.
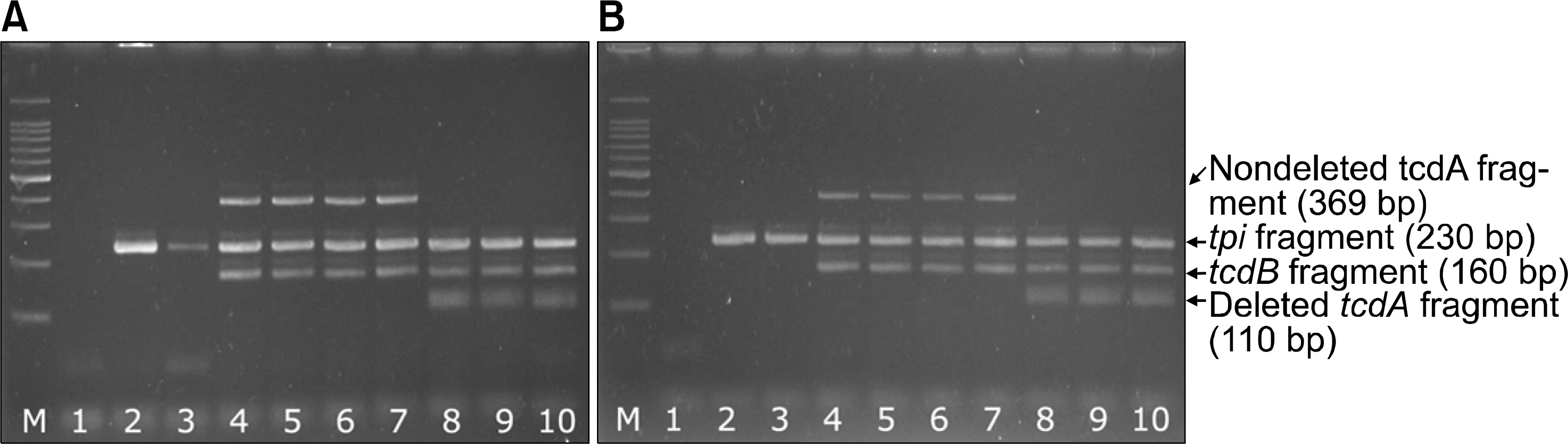
Table 1.
Concordance between multiplex PCR and enzyme immunoassay using VIDAS C. difficile Toxin A&B (CDAB) for toxin production among 29 C. difficile isolates
| CDAB | No. of isolates with toxin profile by multiplex PCR | |||
|---|---|---|---|---|
| A+B+ | A-B+ | A-B- | Total | |
|
Positive Equivocal Negative Total |
5 2 5 12 |
4 2 1 7 |
0 0 10 10 |
9 4 16 29 |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download