초록
Infections and outbreaks of antimicrobial-resistant ba-cteria, such as methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enteroco-ccus (VRE), have been increasing. Detection methods for antimicrobial-resistant bacteria have been changed from traditional culture methods to chromogenic media culture and molecular methods. Strain- typing methods using various molecular technologies are essential tools for epidemiologic surveillance. Furthermore, outbreak detection, using syndromic surveillance as well as passive and active surveillance, has been applied. However, it is difficult to establish effective and robust guidelines and systems for using these various methods to control antimicrobial-resi-stant bacteria. Therefore, clinical microbiologists and policy makers must possess expertise in the control of antimicrobial resistant bacteria, discuss the issue sufficiently, and, finally, create a system to accom-plish this control.
REFERENCES
1.National Nosocomial Infections Surveillance System. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am J Infect Control. 2004. 32:470–85.
2.Obritsch MD., Fish DN., MacLaren R., Jung R. National surveillance of antimicrobial resistance in Pseudomonas aeruginosa isolates obtained from intensive care unit patients from 1993 to 2002. Antimicrob Agents Chemother. 2004. 48:4606–10.
3.Yong D., Toleman MA., Giske CG., Cho HS., Sundman K., Lee K, et al. Characterization of a new metallo-beta-lactamase gene, bla NDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009. 53:5046–54.
4.Hishinuma A., Ishida T. New Delhi metallo-beta-lactamase-1 (NDM-1) producing bacteria. Nihon Rinsho. 2012. 70:262–6.
5.Malhotra-Kumar S., Haccuria K., Michiels M., Ieven M., Poyart C., Hryniewicz W, et al. MOSAR WP2 Study Team. Current trends in rapid diagnostics for methicillin-resistant Staphylococcus aureus and glycopeptide-resistant Enterococcus species. J Clin Microbiol. 2008. 46:1577–87.
6.Ben Nsira S., Dupuis M., Leclercq R. Evaluation of MRSA Select, a new chromogenic medium for the detection of nasal carriage of methicillin-resistant Staphylococcus aureus. Int J Antimicrob Agents. 2006. 27:561–4.
7.Compernolle V., Verschraegen G., Claeys G. Combined use of Pastorex Staph-Plus and either of two new chromogenic agars, MRSA ID and CHROMagar MRSA, for detection of methicillin- resistant Staphylococcus aureus. J Clin Microbiol. 2007. 45:154–8.
8.Louie L., Soares D., Meaney H., Vearncombe M., Simor AE. Evaluation of a new chromogenic medium, MRSA select, for detection of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2006. 44:4561–3.
9.Nahimana I., Francioli P., Blanc DS. Evaluation of three chromogenic media (MRSA-ID, MRSASelect and CHROMagar MRSA) and ORSAB for surveillance cultures of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2006. 12:1168–74.
10.Stoakes L., Reyes R., Daniel J., Lennox G., John MA., Lannigan R, et al. Prospective comparison of a new chromogenic medium, MRSASelect, to CHROMagar MRSA and mannitol-salt medium supplemented with oxacillin or cefoxitin for detection of methi-cillin-resistant Staphylococcus aureus. J Clin Microbiol. 2006. 44:637–9.
11.van Hal SJ., Stark D., Lockwood B., Marriott D., Harkness J. Methicillin-resistant Staphylococcus aureus (MRSA) detection: comparison of two molecular methods (IDI-MRSA PCR assay and GenoType MRSA Direct PCR assay) with three selective MRSA agars (MRSA ID, MRSASelect, and CHROMagar MRSA) for use with infection-control swabs. J Clin Microbiol. 2007. 45:2486–90.
12.Cherkaoui A., Renzi G., François P., Schrenzel J. Comparison of four chromogenic media for culture-based screening of meticillin- resistant Staphylococcus aureus. J Med Microbiol. 2007. 56:500–3.
13.Perry JD., Davies A., Butterworth LA., Hopley AL., Nicholson A., Gould FK. Development and evaluation of a chromogenic agar medium for methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2004. 42:4519–23.
14.Daeschlein G., Assadian O., Daxboeck F., Kramer A. Multiplex PCR-ELISA for direct detection of MRSA in nasal swabs advanta-geous for rapid identification of non-MRSA carriers. Eur J Clin Microbiol Infect Dis. 2006. 25:328–30.

15.Holfelder M., Eigner U., Turnwald AM., Witte W., Weizenegger M., Fahr A. Direct detection of methicillin-resistant Staphylococcus aureus in clinical specimens by a nucleic acid-based hybridisation assay. Clin Microbiol Infect. 2006. 12:1163–7.
16.Levi K., Towner KJ. Rapid detection of methicillin-resistant Staphylococcus aureus from screening enrichment broths by real- time PCR. Eur J Clin Microbiol Infect Dis. 2005. 24:423–7.
17.Paule SM., Hacek DM., Kufner B., Truchon K., Thomson RB Jr., Kaul KL, et al. Performance of the BD GeneOhm methicillin- resistant Staphylococcus aureus test before and during high-volume clinical use. J Clin Microbiol. 2007. 45:2993–8.
18.Wagenvoort JH., van de Cruijs MF., Meuwissen CT., Gronenschild JM., De Brauwer EI. Comparison of an enrichment broth-enhanced commercial PCR procedure versus bacteriological culture for separating non-colonized from suspected or colonized MRSA individuals. Eur J Clin Microbiol Infect Dis. 2007. 26:155–60.

19.Bishop EJ., Grabsch EA., Ballard SA., Mayall B., Xie S., Martin R, et al. Concurrent analysis of nose and groin swab specimens by the IDI-MRSA PCR assay is comparable to analysis by individual- specimen PCR and routine culture assays for detection of colonization by methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2006. 44:2904–8.
20.Rossney AS., Herra CM., Fitzgibbon MM., Morgan PM., Lawrence MJ., O'Connell B. Evaluation of the IDI-MRSA assay on the SmartCycler real-time PCR platform for rapid detection of MRSA from screening specimens. Eur J Clin Microbiol Infect Dis. 2007. 26:459–66.

21.Zhang SX., Drews SJ., Tomassi J., Katz KC. Comparison of two versions of the IDI-MRSA assay using charcoal swabs for prospe-ctive nasal and nonnasal surveillance samples. J Clin Microbiol. 2007. 45:2278–80.

22.Desjardins M., Guibord C., Lalonde B., Toye B., Ramotar K. Evaluation of the IDI-MRSA assay for detection of methicillin- resistant Staphylococcus aureus from nasal and rectal specimens pooled in a selective broth. J Clin Microbiol. 2006. 44:1219–23.
23.Wolk DM., Picton E., Johnson D., Davis T., Pancholi P., Ginocchio CC, et al. Multicenter evaluation of the Cepheid Xpert methicillin- resistant Staphylococcus aureus (MRSA) test as a rapid screening method for detection of MRSA in nares. J Clin Microbiol. 2009. 47:758–64.
24.Domingo MC., Huletsky A., Giroux R., Boissinot K., Picard FJ., Lebel P, et al. High prevalence of glycopeptide resistance genes vanB, vanD, and vanG not associated with enterococci in human fecal flora. Antimicrob Agents Chemother. 2005. 49:4784–6.
25.Li W., Raoult D., Fournier PE. Bacterial strain typing in the genomic era. FEMS Microbiol Rev. 2009. 33:892–916.

26.Struelens MJ., Hawkey PM., French GL., Witte W., Tacconelli E. Laboratory tools and strategies for methicillin-resistant Staphylococcus aureus screening, surveillance and typing: state of the art and unmet needs. Clin Microbiol Infect. 2009. 15:112–9.
27.Carnevale RJ., Talbot TR., Schaffner W., Bloch KC., Daniels TL., Miller RA. Evaluating the utility of syndromic surveillance algori-thms for screening to detect potentially clonal hospital infection outbreaks. J Am Med Inform Assoc. 2011. 18:466–72.

28.Buckeridge DL. Outbreak detection through automated surveillance: a review of the determinants of detection. J Biomed Inform. 2007. 40:370–9.

29.Jernigan JA., Stephens DS., Ashford DA., Omenaca C., Topiel MS., Galbraith M, et al. Anthrax Bioterrorism Investigation Team. Bioterrorism-related inhalational anthrax: the first 10 cases reported in the United States. Emerg Infect Dis. 2001. 7:933–44.

30.Centers for Disease Control and Prevention (CDC). Syndromic surveillance. Reports from a national conference, 2003. MMWR Morb Mortal Wkly Rep. 2004. 53(Suppl):1–264.
31.Centers for Disease Control and Prevention (CDC). Syndromic surveillance for bioterrorism following the attacks on the World Trade Center--New York City, 2001. MMWR Morb Mortal Wkly Rep. 2002. 51:13–5.
32.Begier EM., Sockwell D., Branch LM., Davies-Cole JO., Jones LH., Edwards L, et al. The National Capitol Region's Emergency Department syndromic surveillance system: do chief complaint and discharge diagnosis yield different results? Emerg Infect Dis. 2003. 9:393–6.

33.Gesteland PH., Wagner MM., Chapman WW., Espino JU., Tsui FC., Gardner RM, et al. Rapid deployment of an electronic disease surveillance system in the state of Utah for the 2002 Olympic Winter Games. Proc AMIA Symp. 2002. 285–9.
34.Lazarus R., Kleinman K., Dashevsky I., Adams C., Kludt P., DeMaria A Jr, et al. Use of automated ambulatory-care encounter records for detection of acute illness clusters, including potential bioterrorism events. Emerg Infect Dis. 2002. 8:753–60.

35.Wagner MM., Tsui FC., Espino JU., Dato VM., Sittig DF., Caruana RA, et al. The emerging science of very early detection of disease outbreaks. J Public Health Manag Pract. 2001. 7:51–9.

36.Kenner J., O'Connor T., Piantanida N., Fishbain J., Eberly B., Viscount H, et al. Rates of carriage of methicillin-resistant and methicillin-susceptible Staphylococcus aureus in an outpatient po-pulation. Infect Control Hosp Epidemiol. 2003. 24:439–44.
37.van den Braak N., Ott A., van Belkum A., Kluytmans JA., Koeleman JG., Spanjaard L, et al. Prevalence and determinants of fecal colonization with vancomycin-resistant Enterococcus in hospitalized patients in The Netherlands. Infect Control Hosp Epidemiol. 2000. 21:520–4.
38.Coello R., Glynn JR., Gaspar C., Picazo JJ., Fereres J. Risk factors for developing clinical infection with methicillin-resistant Staphylococcus aureus (MRSA) amongst hospital patients initially only colonized with MRSA. J Hosp Infect. 1997. 37:39–46.
39.Huang SS., Platt R. Risk of methicillin-resistant Staphylococcus aureus infection after previous infection or colonization. Clin Infect Dis. 2003. 36:281–5.
40.Pujol M., Peña C., Pallares R., Ayats J., Ariza J., Gudiol F. Risk factors for nosocomial bacteremia due to methicillin-resistant Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. 1994. 13:96–102.
41.Henning KJ., Delencastre H., Eagan J., Boone N., Brown A., Chung M, et al. Vancomycin-resistant Enterococcus faecium on a pediatric oncology ward: duration of stool shedding and incidence of clinical infection. Pediatr Infect Dis J. 1996. 15:848–54.
42.Yeh KM., Siu LK., Chang JC., Chang FY. Vancomycin-resistant enterococcus (VRE) carriage and infection in intensive care units. Microb Drug Resist. 2004. 10:177–83.
43.Zirakzadeh A., Patel R. Vancomycin-resistant enterococci: colonization, infection, detection, and treatment. Mayo Clin Proc. 2006. 81:529–36.

44.Pittet D., Hugonnet S., Harbarth S., Mourouga P., Sauvan V., Touveneau S, et al. Effectiveness of a hospital-wide programme to improve compliance with hand hygiene. Infection Control Progra-mme. Lancet. 2000. 356:1307–12.
45.Johnson PD., Martin R., Burrell LJ., Grabsch EA., Kirsa SW., O'Keeffe J, et al. Efficacy of an alcohol/chlorhexidine hand hygiene program in a hospital with high rates of nosocomial methicillin-resistant Staphylococcus aureus (MRSA) infection. Med J Aust. 2005. 183:509–14.
46.Rosenthal VD., Guzman S., Safdar N. Reduction in nosocomial infection with improved hand hygiene in intensive care units of a tertiary care hospital in Argentina. Am J Infect Control. 2005. 33:392–7.

47.Dancer SJ., Coyne M., Speekenbrink A., Samavedam S., Kennedy J., Wallace PG. MRSA acquisition in an intensive care unit. Am J Infect Control. 2006. 34:10–7.

48.Jernigan JA., Titus MG., Gröschel DH., Getchell-White S., Farr BM. Effectiveness of contact isolation during a hospital outbreak of methicillin-resistant Staphylococcus aureus. Am J Epidemiol. 1996. 143:496–504.
49.Puzniak LA., Leet T., Mayfield J., Kollef M., Mundy LM. To gown or not to gown: the effect on acquisition of vancomycin-resistant enterococci. Clin Infect Dis. 2002. 35:18–25.

50.Karanfil LV., Murphy M., Josephson A., Gaynes R., Mandel L., Hill BC, et al. A cluster of vancomycin-resistant Enterococcus faecium in an intensive care unit. Infect Control Hosp Epidemiol. 1992. 13:195–200.
51.Montecalvo MA., Horowitz H., Gedris C., Carbonaro C., Tenover FC., Issah A, et al. Outbreak of vancomycin-, ampicillin-, and amino-glycoside-resistant Enterococcus faecium bacteremia in an adult oncology unit. Antimicrob Agents Chemother. 1994. 38:1363–7.
52.Back NA., Linnemann CC Jr., Staneck JL., Kotagal UR. Control of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit: use of intensive microbiologic surveillance and mupirocin. Infect Control Hosp Epidemiol. 1996. 17:227–31.
53.Armstrong-Evans M., Litt M., McArthur MA., Willey B., Cann D., Liska S, et al. Control of transmission of vancomycin-resistant Enterococcus faecium in a long-term-care facility. Infect Control Hosp Epidemiol. 1999. 20:312–7.
54.Byers KE., Anglim AM., Anneski CJ., Germanson TP., Gold HS., Durbin LJ, et al. A hospital epidemic of vancomycin-resistant Enterococcus: risk factors and control. Infect Control Hosp Epidemiol. 2001. 22:140–7.
55.Saiman L., Cronquist A., Wu F., Zhou J., Rubenstein D., Eisner W, et al. An outbreak of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit. Infect Control Hosp Epidemiol. 2003. 24:317–21.
56.Khoury J., Jones M., Grim A., Dunne WM Jr., Fraser V. Eradication of methicillin-resistant Staphylococcus aureus from a neonatal intensive care unit by active surveillance and aggressive infection control measures. Infect Control Hosp Epidemiol. 2005. 26:616–21.
57.Singh N., Léger MM., Campbell J., Short B., Campos JM. Control of vancomycin-resistant enterococci in the neonatal intensive care unit. Infect Control Hosp Epidemiol. 2005. 26:646–9.

58.Mascini EM., Troelstra A., Beitsma M., Blok HE., Jalink KP., Hopmans TE, et al. Genotyping and preemptive isolation to control an outbreak of vancomycin-resistant Enterococcus faecium. Clin Infect Dis. 2006. 42:739–46.
59.Huskins WC., Huckabee CM., O'Grady NP., Murray P., Kopetskie H., Zimmer L, et al. STAR*ICU Trial Investigators. Intervention to reduce transmission of resistant bacteria in intensive care. N Engl J Med. 2011. 364:1407–18.

60.Dembry LM., Uzokwe K., Zervos MJ. Control of endemic glycopeptide-resistant enterococci. Infect Control Hosp Epidemiol. 1996. 17:286–92.

61.Jochimsen EM., Fish L., Manning K., Young S., Singer DA., Baker R, et al. Control of vancomycin-resistant enterococci at a community hospital: efficacy of patient and staff cohorting. Infect Control Hosp Epidemiol. 1999. 20:106–9.

62.Siddiqui AH., Harris AD., Hebden J., Wilson PD., Morris JG Jr., Roghmann MC. The effect of active surveillance for vancomycin- resistant enterococci in high-risk units on vancomycin-resistant enterococci incidence hospital-wide. Am J Infect Control. 2002. 30:40–3.
63.Huang SS., Yokoe DS., Hinrichsen VL., Spurchise LS., Datta R., Miroshnik I, et al. Impact of routine intensive care unit surveillance cultures and resultant barrier precautions on hospital-wide methi-cillin-resistant Staphylococcus aureus bacteremia. Clin Infect Dis. 2006. 43:971–8.
64.Karchmer TB., Durbin LJ., Simonton BM., Farr BM. Cost-effecti-veness of active surveillance cultures and contact/droplet precautions for control of methicillin-resistant Staphylococcus aureus. J Hosp Infect. 2002. 51:126–32.
65.Weber SG., Huang SS., Oriola S., Huskins WC., Noskin GA. $$$$.
Fig. 1.
Syndromic surveillance: rationale for early detection. *Time between detection by syndromic (prediagnostic) surveillance and detection by traditional (diagnosis-bases) surveillance. Figure is adapted from MMWR, 2004;53.
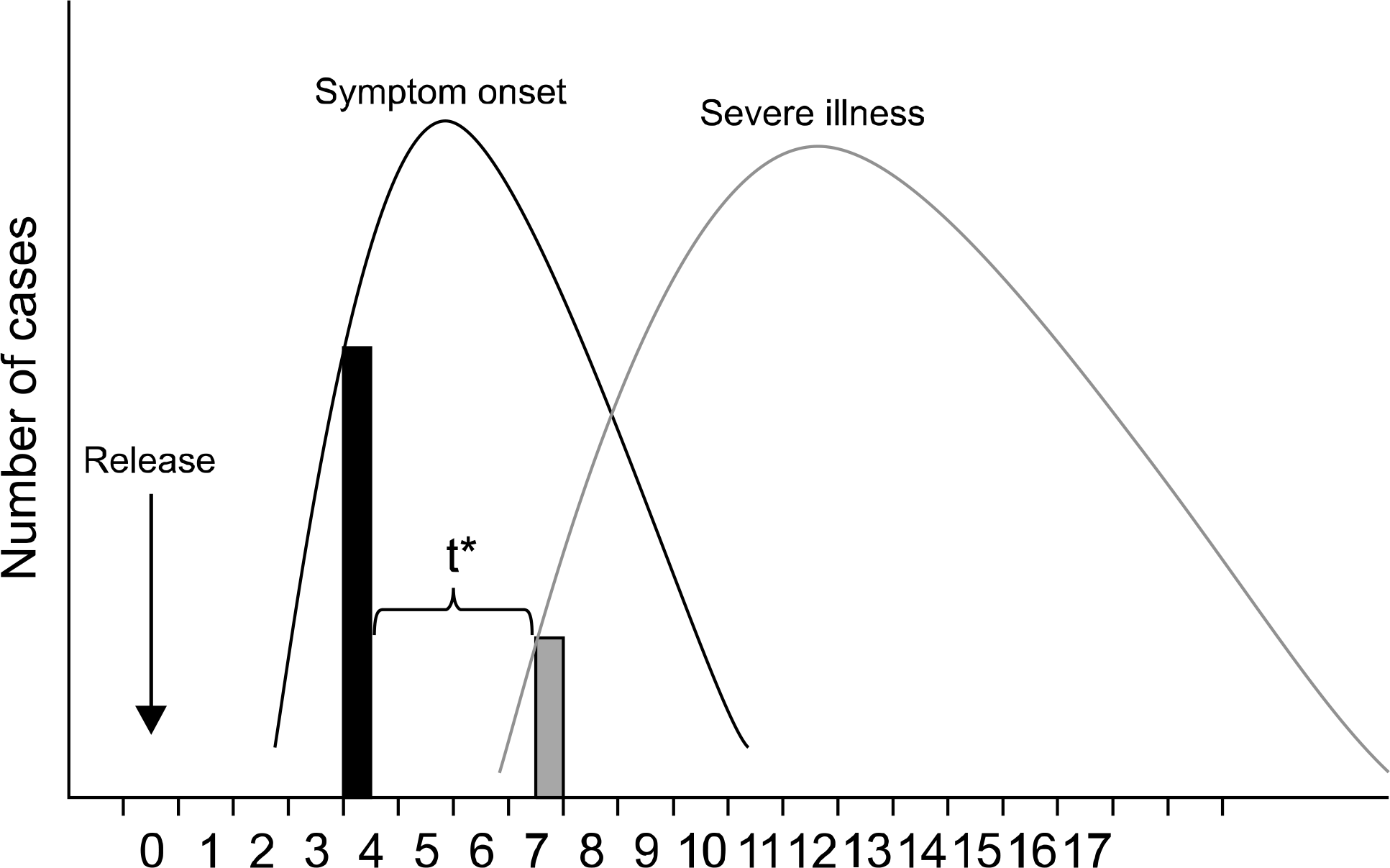
Fig. 2.
Legislative maps of methicillin-resistant Staphylococcus aureus in the United States. Figure is adapted from http://apic.org.
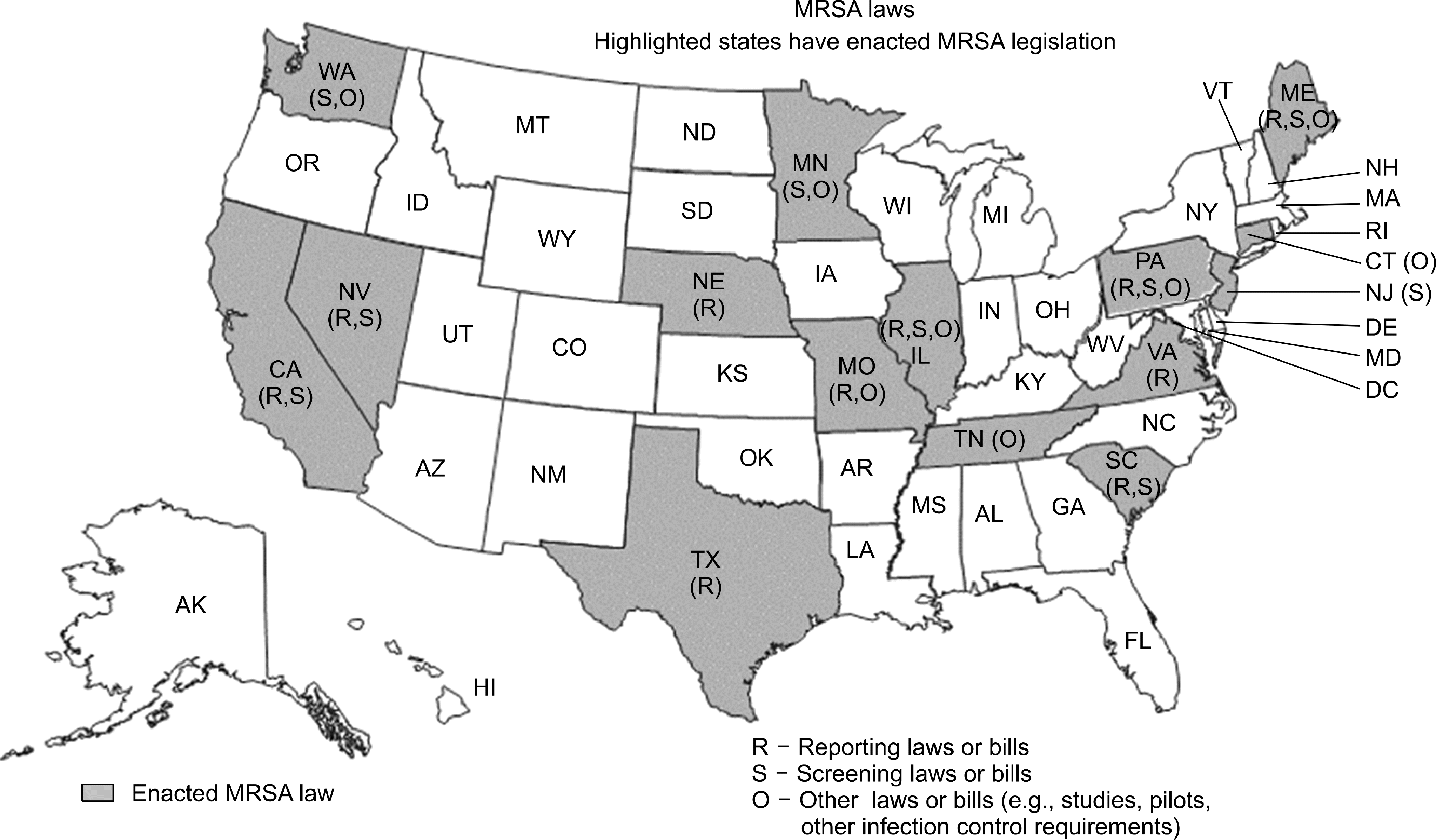
Table 1.
Sensitivity and specificity of currently available chromogenic media and molecular assays for methicillin-resistant Staphylococcus aureus
Table 2.
Required elements of an effective active surveillance program




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download