TO THE EDITOR: According to the 2008 World Health Organization (WHO) guidelines, eosinophilia is associated with hematologic diseases such as chronic eosinophilic leukemia-not otherwise specified (CEL-NOS); idiopathic hypereosinophilic syndrome (HES) or idiopathic hypereosinophilia; and myeloid and lymphoid neoplasms with eosinophilia, and abnormalities of PDGFRA, PDGFRB or FGFR1 [1]. CEL is diagnosed in cases with increased blasts or cytogenetic/clonal abnormalities. We report 2 cases of myeloid and lymphoid neoplasms (CEL) with the FIP1L1-PDGFRA rearrangement.
The first case, 27-year-old man was referred to our hospital for a dyspnea workup. The echocardiogram revealed severe tricuspid valve regurgitation and moderate mitral valve regurgitation. A valvuloplasty was performed and dizziness developed after the surgery. Brain magnetic resonance imaging revealed microbleeding in the right temporo-occipital lobe. He was referred to the hemato-oncology department for evaluation of eosinophilia. The laboratory findings were as follows: WBC count, 21.1×109/L (eosinophil count, 13.0×109/L); Hb, 9.1 g/dL; and platelet count, 99.0×109/L. In addition, parasite antibodies for Clonorchis sinensis, Paragonimus westermani, Cysticercus cellulosae, and Sparganosis were negative. The number of eosinophils in the peripheral blood smear was increased (Fig. 1A). Analysis of the bone marrow aspirate showed that 1.95% of all nucleated cells were blasts and eosinophils were increased (Fig. 1B). Conventional cytogenetic analysis revealed a normal karyotype (46,XY). Fluorescence in situ hybridization (FISH) for FIP1L1-PDGFRA using the Vysis 4q12 Tri-Color probe (Abbott, Des Plaines, USA) was positive (Fig. 2A, B). We confirmed the FIP1L1-PDGFRA rearrangement by reverse transcription-polymerase chain reaction (RT-PCR) and sequencing analysis (Fig. 2D, E). The primers for RT-PCR and sequencing analysis were as follows: forward, 5'-TCAGACAAGTACTGCCTCCAGA-3' and reverse, 5'-AG GCTCCCAGCAAGTTTACA-3'. According to the 2008 WHO guidelines, a diagnosis of myeloid and lymphoid neoplasm (CEL) associated with FIP1L1-PDGFRA was established. Tyrosine kinase inhibitor treatment with imatinib mesylate was initiated and the eosinophil counts decreased.
The second case, 23-year-old man was referred to our hospital for a workup as eosinophilia had been detected in a routine health checkup. The laboratory findings were as follows: WBC count, 70.0×109/µL (eosinophil count, 55.3×109/L); Hb, 10.1 g/dL; and platelet count, 131.0×109/L. The number of eosinophils in the peripheral blood smear was increased (Fig. 1C). In the bone marrow aspirate, 90% of all nucleated cells were eosinophils. Infiltration by eosinophils was observed in the bone marrow section (Fig. 1D). Conventional cytogenetic analysis revealed a normal karyotype (46,XY) and FISH for FIP1L1-PDGFRA was positive (Fig. 2C). Tyrosine kinase inhibitor treatment with imatinib mesylate was initiated and the eosinophil counts decreased.
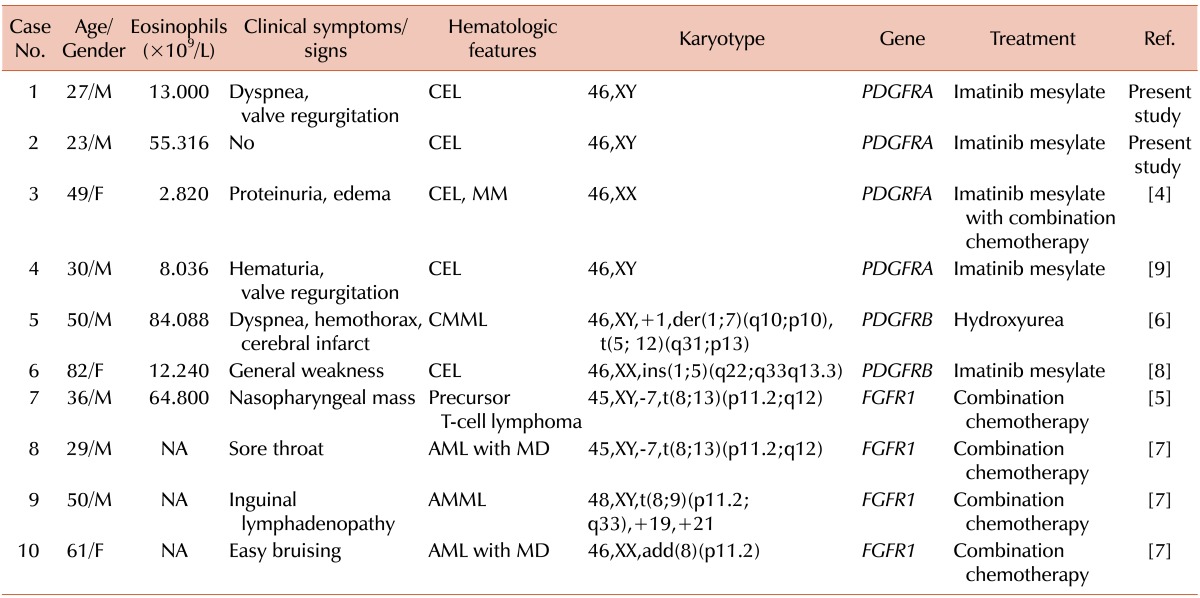
The FIP1L1-PDGFRA rearrangement caused by a cryptic deletion of 800-kb on chromosome 4q12, which contains the CHIC2 gene [2], cannot be detected in conventional chromosomal studies; FISH or RT-PCR must be used. The prevalence of PDGFRA, PDGFRB or FGFR1 rearrangements is generally low. The FIP1L1-PDGFRA rearrangement is the most commonly found with an incidence of approximately 23%, but this varies (range, 3-56%) depending on the HES patient population [3], and is predominantly found in males. In a Korean study [4], only 1 case with a PDGFRA rearrangement was detected among 34 hypereosinophilia patients who were suspected of having clonal eosinophilia. Until now, few cases of PDGFRA, PDGFRB, or FGFR1 rearrangements have been reported in Korea [4, 5, 6, 7, 8, 9], and these are summarized in Table 1.
The initial clinical manifestation of HES involves dermatologic, pulmonary, and gastrointestinal symptoms [10]. However, the presence of neurologic or cardiac signs and symptoms increases with disease progression or delayed diagnosis [10]. Severe cardiac complications include endomyocardial fibrosis, valve scarring, and embolism. Our patients also showed dyspnea, cardiac signs (valve regurgitation), and renal involvement (Case 1). Therefore, clinicians need to be alert for the presence PDGFRA, PDGFRB, or FGFR1 rearrangements in eosinophilia patients with systemic symptoms, especially those associated with the cardiopulmonary or renal systems.
In conclusion, we report 2 cases of CEL with the FIP1L1-PDGFRA rearrangement. Eosinophilia with PDGFRA, PDGFRB, or FGFR1 rearrangements is rare in the general population. However, its prevalence is high in HES patients (range, 3-56%) [3], and FISH or RT-PCR must be used for its detection. Unusual cardiopulmonary or gastrointestinal symptoms with severe complications may be observed initially. Therefore, clinicians must consider PDGFRA, PDGFRB or FGFR1 rearrangements in patients with eosinophilia presenting with these symptoms and perform FISH or RT-PCR.
References
1. Swerdlow SH, Campo E, Harris NL, editors. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th ed. Lyon, France: IARC Press;2008.
2. Pardanani A, Ketterling RP, Brockman SR, et al. CHIC2 deletion, a surrogate for FIP1L1-PDGFRA fusion, occurs in systemic mastocytosis associated with eosinophilia and predicts response to imatinib mesylate therapy. Blood. 2003; 102:3093–3096. PMID: 12842979.

3. Gotlib J, Cools J. Five years since the discovery of FIP1L1-PDGFRA: what we have learned about the fusion and other molecularly defined eosinophilias. Leukemia. 2008; 22:1999–2010. PMID: 18843283.

4. Kim DW, Shin MG, Yun HK, et al. Incidence and causes of hypereosinophilia (corrected) in the patients of a university hospital. Korean J Lab Med. 2009; 29:185–193. PMID: 19571614.
5. Park TS, Song J, Kim JS, et al. 8p11 myeloproliferative syndrome preceded by t(8;9)(p11;q33), CEP110/FGFR1 fusion transcript: morphologic, molecular, and cytogenetic characterization of myeloid neoplasms associated with eosinophilia and FGFR1 abnormality. Cancer Genet Cytogenet. 2008; 181:93–99. PMID: 18295660.

6. Kim M, Lim J, Lee A, et al. A case of chronic myelomonocytic leukemia with severe eosinophilia having t(5;12)(q31;p13) with t(1;7)(q10;p10). Acta Haematol. 2005; 114:104–107. PMID: 16103634.

7. Lee H, Kim M, Lim J, et al. Acute myeloid leukemia associated with FGFR1 abnormalities. Int J Hematol. 2013; 97:808–812. PMID: 23609419.

8. Jang SE, Kang HJ, Chang YH, et al. A case of myeloid neoplasm with the PDGFRB rearrangement and eosinophilia. Korean J Med. 2010; 78:386–390.
9. Lim KS, Ko J, Lee SS, Shin B, Choi DC, Lee BJ. A case of idiopathic hypereosinophilic syndrome presenting with acute respiratory distress syndrome. Allergy Asthma Immunol Res. 2014; 6:98–101. PMID: 24404401.

10. Ogbogu PU, Bochner BS, Butterfield JH, et al. Hypereosinophilic syndrome: a multicenter, retrospective analysis of clinical characteristics and response to therapy. J Allergy Clin Immunol. 2009; 124:1319–1325. PMID: 19910029.

Fig. 1
Peripheral blood (PB) smear and bone marrow (BM) aspirate findings of first case (A, B) and second case (C, D). (A) Eosinophilia (13.0×109/L) was observed in the PB (Wright-Giemsa, ×1,000). (B) Dysplastic eosinophils (sparse and mixed granules) were increased in the BM aspirate (Wright-Giemsa, ×1,000). (C) Eosinophilia (55.316×109/L) was observed in the PB (Wright-Giemsa, ×1,000). (D) The BM section was packed with eosinophils (hematoxylin-eosin, ×400).
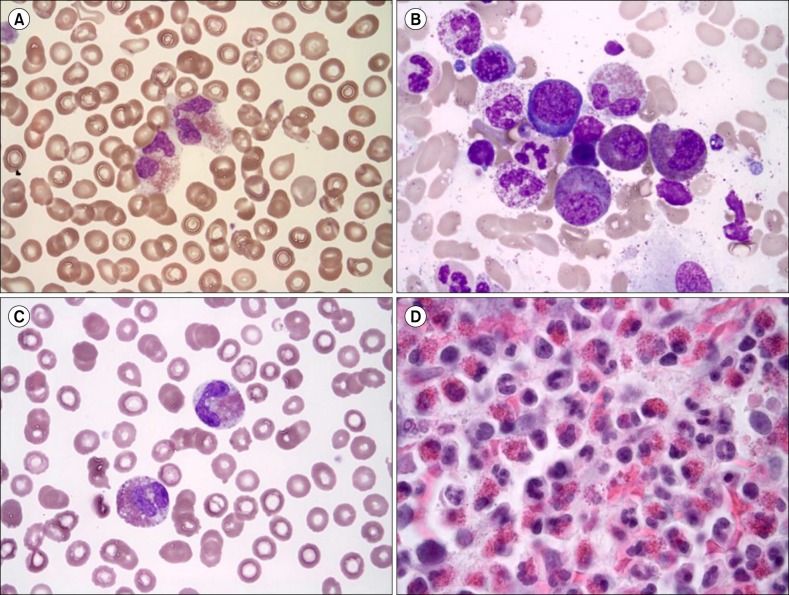
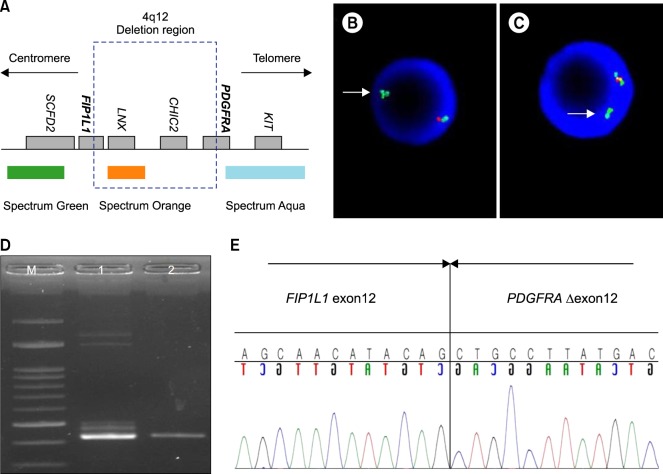
Fig. 2
Fluorescence in situ hybridization (FISH) for the FIP1L1-PDGFRA rearrangement, and reverse transcription - polymerase chain reaction (RT-PCR) and sequencing analysis. (A) Schematic representation of the 4q12 region and FISH probe. (B) Loss of the orange signal indicates deletion of the 4q12 region (white arrow) in first case. (C) FISH results for second case. (D) RT-PCR was performed using RNA extracted from the patient's (first case) white blood cells (lane 1). EOL-1 cells were used as a positive control (lane 2). Size marker (M). (E) Sequencing analysis of the RT-PCR products revealed the fusion of FIP1L1, exon 12 and truncated PDGFRA, exon 12 (breakpoint is between the 84th and 85th nucleotides of exon 12). FIP1L1, Ensemble Gene ID ENSG00000145216; PDGFRA, Ensemble Gene ID, ENSG00000134853.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download