Abstract
Background
Malassezia species play an important role in the pathogenesis of seborrheic dermatitis. In particular, M. restricta and M. globosa are considered to be the predominant organisms in seborrheic dermatitis of Western countries. However, species distribution of Malassezia in seborrheic dermatitis has not been clearly determined yet in Asia.
Objective
To identify the distribution of Malassezia species on the scalp of seborrheic dermatitis patients in Korea using 26S rDNA PCR-RFLP analysis.
Methods
A total of 40 seborrheic dermatitis patients and 100 normal healthy volunteers were included in this study. For the identification of Malassezia species, the scalp scales of the subjects were analyzed by 26S rDNA PCR-RFLP analysis.
Results
The most commonly identified Malassezia species were M. restricta in the seborrheic dermatitis patients, and M. globosa in the normal controls. In the seborrheic dermatitis group, M. restricta was identified in 47.5%, M. globosa in 27.5%, M. furfur in 7.5%, and M. sympodialis in 2.5% of patients. In the healthy control group, M. globosa was identified in 32.0%, M. restricta in 25.0%, M. furfur in 8.0%, M. obtusa in 6.0%, M. slooffiae in 6.0%, and M. sympodialis in 4.0% of subjects.
Malassezia species are lipophilic fungi which are regarded as part of the normal flora of human skin, and are found in 75~98% of healthy adults. These yeasts are the cause of pityriasis versicolor and Malassezia folliculitis, and appear to be involved in the pathogenesis of common skin disorders such as seborrheic dermatitis, psoriasis, and atopic dermatitis1. In 1996, Guého et al.2 classified Malassezia into seven species, M. furfur, M. obtusa, M. globosa, M. slooffiae, M. sympodialis, M. pachydermatis, and M. restricta, based on morphological and biochemical analysis. However, with the development of molecular methods such as polymerase chain reaction (PCR), an additional six species have been described which are M. yamatoensis, M. nana, M. japonica, M. equine, M. caprae and M. dermatis3. Among them, M. restricta and M. globosa are considered to be the most important pathogenic organisms in the development of seborrheic dermatitis4. However, some reports have also linked M. furfur, M. sympodialis, M. obtusa, and M. slooffiae with seborrheic dermatitis5.
The aim of this study was to identify Malassezia species on the scalp of 40 patients with seborrheic dermatitis in Korea using 26S rDNA polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP). We also compared the results of seborrheic dermatitis patients with those of 100 normal healthy volunteers.
Six reference strains of Malassezia species, which are known to be involved in the pathogenesis of seborrheic dermatitis, were used to determine the 26S rDNA PCR-RFLP pattern of Malassezia yeasts in this study4,5. The standard strains were purchased from Centralbureau Boor Schimmelcultures (CBS, Utrecht, Netherlands), Biological Resource Center and included M. furfur (CBS 1878), M. restricta (CBS 7877), M. obtusa (CBS 7876), M. sympodialis (CBS 7222), M. slooffiae (CBS 7956) and M. globosa (CBS 7966).
A total of 140 subjects, including 40 seborrheic dermatitis (with scalp involvement) patients and 100 healthy volunteers as controls, were enrolled in this study (Table 1). Seborrheic dermatitis on the lesions of the scalp was clinically diagnosed, with seborrhea, greasy, yellowish, pityriasiform scaling related to irregular sharply delimited erythematous plaques. Some seborrheic dermatitis patients had pronounced seborrhea and inflamed erythematous folliculitis. The patients without scalp lesions or with only dandruff on the scalp lesions were excluded. We included only those patients who had not used any topical or oral treatment during the previous 2 months. No concomitant diseases were registered. The mean age was significantly higher in the seborrheic dermatitis group (42.5, range 25~54) than in the control group (35.7, range 22~66) (p=0.001). The proportion of males was significantly higher in the seborrheic dermatitis group (65.0%) than in the control group (60.0%) (p=0.045).
Scalp scales were collected by swabbing in both the seborrheic dermatitis and normal control group. A sterile cotton swab was moistened in 100µl of the detergent fluid containing 0.1% Triton X-100 in 0.075 mol phosphate buffer (pH 7.9), and was rubbed five times against a round area of 3 cm diameter on the scalp skin. Then, only the fiber-tipped part of the swab was cut and placed in 900µl of the detergent fluid. This was placed into a shaker for 30 seconds to evenly disperse the Malassezia yeasts in the solution. Approximately 100µl of sampled skin was mixed with 900µl of the detergent. From this, 100µl was taken and applied evenly on Leeming & Notman culture medium, and incubated at 34℃ for 14 days.
Genomic DNA isolation was perfomed by the glass beads method. The obtained Malassezia yeasts from the patients were suspended in 400µl lysis buffer (100 mM Tris-HCl pH 8.0, 1.0% SDS, 2.0% Triton X-100, 10 mM EDTA, 100 mM NaCl). A total of 400µl P/C/I (phenol: chloroform: isoamyl alcohol=25:24:1, v/v) and 400 mg glass beads (diameter 0.5 mm) were added. This mixture was vortexed for 10 minutes and centrifuged at 15,000 rpm for 10 minutes. Total DNA was precipitated from the aqueous fraction with isopropanol and centrifuged at 15,000 rpm for 15 minutes. The DNA pellet was washed in 70% ethanol and resuspended in sterile water.
Primers were designed to amplify the 26S rDNA region of all six species of Malassezia. The sequences were forward, 5'-TAACAAGGATTCCCCTAGTA-3' and reverse, 5'-ATTACGCCAGCATCCTAAG-3'. The conditions of the early stage of reaction were as follows: pre-denaturation at 95℃ for 14 minutes, denaturation at 94℃ for 45 seconds, annealing at 55℃ for 45 seconds, extension of 35 cycles at 72℃ for 1 minute, and final extension at 72℃ for 7 minutes. Amplified DNA was confirmed by electrophoresis on a 1.5% agarose gel stained with ethidium bromide (0.5µg/ml) by using 1 × TAE migrating buffer (pH 8.0, 40 mM Tri-acetate 1 mM EDTA).
The amplified 26S rDNA was purified using the Accu-Prep PCR purification kit (Bioneer, Daejeon, Korea). Two restriction enzymes, Hha I (Takara Biomedicals, Otsu, Japan) and BstF51 (SibEnzyme, Novosibirsk, Russia) were used. The restriction enzyme reaction was performed with 10 × PCR buffer, 10 U of the restriction enzyme, and 7.5µl of the PCR products. After the reaction, the RFLP pattern was analyzed with DNA fragments in gel electrophoresis.
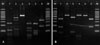
Using colony PCR to amplify the 26S rDNA of Malassezia standard strains, 580 bp PCR bands were confirmed in all of the six standard strains including M. furfur, M. globosa, M. restricta, M. obtusa, M. sympodialis, and M. slooffiae (Fig. 1). All the six standard strains showed different RFLP patterns using the restriction enzyme Hha1. With the restriction enzyme BstF51, M. globosa and M. obtusa showed the same RFLP pattern, whereas the other four strains showed different patterns (Fig. 2). Therefore, we confirmed that these six standard strains can be identified and differentiated by 26S rDNA PCR-RFLP analysis.
Malassezia yeasts of the seborrheic dermatitis group were cultured in 34 out of 40 specimens, (identification rate 85%). In the seborrheic dermatitis group, M. restricta (47.5%) was most commonly recovered, and M. globosa (27.5%) was the second most commonly recovered species. M. furfur and M. sympodialis were identified in 7.5% and 2.5% of the seborrheic dermatitis patients, respectively. M. obtusa and M. slooffiae were not recovered in any of the seborrheic dermatitis patients.
Malassezia yeasts of the control group were cultured in 81 out of 100 specimens, and thus the identification rate was 81%. In the control group, M. globosa (32.0%) was most commonly recovered, and M. restricta (25.0%) was the second most commonly recovered species. The complete set of results for the recovered species is shown in Table 2. Only the most abundant species for each individual was counted, not accounting for the combined infection of two or more species.
In comparison to the seborrheic dermatitis group with the control group, the prevalence of M. restricta was significantly higher in the seborrheic dermatitis group (47.5%) than in the control group (25.0%) (p<0.05). The other five species, M. furfur, M. globosa, M. obtusa, M. sympodialis, and M. slooffiae, did not show the significant differences in prevalence between the seborrheic dermatitis group and the control group.
The prevalence of co-colonization of two or more Malassezia species was 15.0% in the seborrheic dermatitis group, and slightly higher in the control group, at 21.0%. This difference was not statistically significant (p=0.339). The recovered species are shown in Table 3. Co-colonization of M. restricta with M. globosa was the most common in both groups.
The identified Malassezia species did not show significant differences among the different age groups (20~29, 30~39, ≥40) both in the seborrheic dermatitis and control group. According to sex, Malassezia species neither showed significant differences in either the seborrheic dermatitis or control group.
Recently, various molecular biological methods have been applied for the identification of Malassezia species, such as nested PCR, real-time PCR, RFLP, and sequencing analysis6-13. 26S rDNA PCR-RFLP is one such method that uses 26S rDNA containing highly conserved sequences yet enough sequence variation for specific interspecies identification. This method requires only PCR and one or two restriction enzymes. Therefore, it is less demanding than most the other molecular biological approaches13. There are five reports of the successful use of 26S rDNA PCR-RFLP for the identification of Malassezia species13-17. In this study, we also confirmed the accuracy and effectiveness of this method.
Seborrheic dermatitis is a chronic skin disease affecting the seborrheic areas of the body including the scalp, face, and upper trunk. It manifests as erythema, scales, and follicular papules. In seborrheic dermatitis patients, Malassezia yeasts show a significantly higher count and density on the lesional skin than on the normal skin of the same patient and on the skin of healthy controls4. The yeast density is considered the basic difference between normal skin and skin with seborrheic dermatitis. However, this does not correlate positively with the severity of the disease4,18. In previous reports, the species distribution of Malassezia in seborrheic dermatitis showed geographical peculiarities. In Japan and USA, M. globosa was most frequently isolated in seborrheic dermatitis, whereas M. restricta was the second most frequently isolated. In Eastern Europe, M. obtusa was the most frequently recovered species4. As for the species distribution of Malassezia in Korean seborrheic dermatitis patients, there has been only one report by Lim et al.19. In this report, M. restricta was most commonly identified, in eight of 10 patients, and M. globosa was recovered in only one patient. In our study, 34 Korean seborrheic dermatitis patients were investigated, and M. restricta was also most commonly identified (55.9%), showing a significantly higher recovery rate compared to the normal controls (30.9%). However, Jang et al.13 recently reported M. restricta was identified in 56.8% of healthy Korean scalp skin samples. Their results also showed a higher prevalence of M. restricta in older age groups, in contrast to a higher prevalence of M. globosa in younger age groups. Because our seborrheic dermatitis patients had a higher age than the healthy controls, we cannot completely exclude that the age difference between the two groups may act as a confounding factor.
Age may affect the prevalence of Malassezia infection, because Malassezia species are lipophilic fungi, and therefore, the growth of Malassezia greatly depends on the sebum content which differs according to age. The recovery rate of Malassezia is known to be highest in the twenties when sebaceous glands show greatest activity20. However, there have not been many reports regarding the age-dependent difference of Malassezia species in seborrheic dermatitis. Jang et al.13 reported the results of healthy individuals, not of seborrheic dermatitis patients, which showed predominance of M. globosa in the younger age groups and M. restricta in older age groups. However, in another study with 770 healthy Japanese, M. restricta predominated in younger ages, whereas M. globosa predominated in older ages, which is inverse to the Korean report13,20. We can confirm that the distribution of Malassezia species is greatly influenced by geographical locations or ethnic groups. In our study, identified Malassezia species did not differ significantly according to age groups. This could be due to the relatively small number of the subjects and the narrow range of the participants' age in our study.
As for the gender-dependent difference of Malassezia infection in seborrheic dermatitis, there have been no reports yet. In our study, there was no gender-dependent difference in the distribution of Malassezia species in the seborrheic dermatitis or control group. As for healthy skin, there have been a few reports regarding the genderdependent difference of Malassezia distribution13,20. In a Japanese study, M. globosa to M. restricta ratio changed noticeably after 16 years in males, and 23 years in females20.
In our study, 15.0% of seborrheic dermatitis patients showed co-colonization of two or more Malassezia species. This was similar to the previous report by Gaitanis et al.21, which showed that 18% of Greek seborrheic dermatitis patients were co-infected with M. globosa, the most commonly identified species in Greeks, together with other Malassezia species21. However, as a higher prevalence of co-colonization was found in our healthy volunteers (21.0%), co-colonization of two or more Malassezia species may not be important in the development of seborrheic dermatitis.
In our study, only the six reference strains of Malassezia species, which have been reported to be related to seborrheic dermatitis, were used, and the newly reported species, including M. yamatoensis, M. nana, M. japonica, M. equine, M. caprae and M.dermatis, were not included. As there have been a few reports regarding the roles of these new species in seborrheic dermatitis, further studies will be important.
This study is a culture-based study on data gathered from Korean subjects to examine distribution of Malassezia yeast. Culture-based techniques are expected to be useful in the clinical assessment of Malassezia species associated with other fungal infections. However, this conventional method was complex in culture conditions and had a lower chance of successful culture. The assessment of additional types of clinical samples is indicated to determine the broad applicability of the approach. Recently, non-culture based techniques which avoided the culture process and directly extracted fungal DNA and differentiated Malassezia species was introduced22,23. Further studies are required to evaluate this new technique and compare the differences in the rate of successful identification on the scalp in seborrheic dermatitis patients.
In conclusion, M. restricta appears to be the most important Malassezia species in Korean seborrheic dermatitis patients. Age and gender do not seem to greatly influence the distribution of Malassezia species. This was a pioneer study which first investigated more than ten Korean seborrheic dermatitis patients. Further studies including a larger numbers of patients will be needed to confirm our results.
Figures and Tables
Fig. 1
26S rDNA PCR in the six Malassezia standard strains. The size of PCR products were almost 580 bp. Lane M: marker, lane 1: M. furfur, lane 2: M. globosa, lane 3: M. restricta, lane 4: M. obtusa, lane 5: M. sympodialis, lane 6: M. slooffiae.
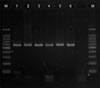
Fig. 2
26S rDNA PCR-RFLP pattern in the six Malassezia standard strains. 26S rDNA PCR products were digested with two restriction enzymes. (A) Hha1; all the six standard strains showed different RFLP patterns. (B) BstF51; M. globosa and M. obtusa showed the same RFLP patterns. The other four strains all showed different patterns. Lane M: marker, lane 1: M. furfur, lane 2: M. globosa, lane 3: M. restricta, lane 4: M. obtusa, lane 5: M. sympodialis, lane 6: M. slooffiae.
References
1. Janik MP, Heffernan MP. Wolff K, Goldsmith LA, Katz SI, Gilchrest BA, Paller AS, Leffell DJ, editors. Yeast infections: candidiasis, pityriasis versicolor. Fitzpatrick's dermatology in general medicine. 2008. 7th ed. New York: McGraw-Hill;1822–1830.
2. Guého E, Midgley G, Guillot J. The genus Malassezia with description of four new species. Antonie Van Leeuwenhoek. 1996. 69:337–355.

3. Sugita T, Tajima M, Ito T, Saito M, Tsuboi R, Nishikawa A. Antifungal activities of tacrolimus and azole agents against the eleven currently accepted Malassezia species. J Clin Microbiol. 2005. 43:2824–2829.

4. Zisova LG. Malassezia species and seborrheic dermatitis. Folia Med (Plovdiv). 2009. 51:23–33.
5. Gupta AK, Batra R, Bluhm R, Boekhout T, Dawson TL Jr. Skin diseases associated with Malassezia species. J Am Acad Dermatol. 2004. 51:785–798.
6. Morishita N, Sei Y, Sugita T. Molecular analysis of Malassezia microflora from patients with pityriasis versicolor. Mycopathologia. 2006. 161:61–65.

7. Sugita T, Tajima M, Tsubuku H, Tsuboi R, Nishikawa A. Quantitative analysis of cutaneous Malassezia in atopic dermatitis patients using real-time PCR. Microbiol Immunol. 2006. 50:549–552.

8. Gupta AK, Boekhout T, Theelen B, Summerbell R, Batra R. Identification and typing of Malassezia species by amplified fragment length polymorphism and sequence analyses of the internal transcribed spacer and large-subunit regions of ribosomal DNA. J Clin Microbiol. 2004. 42:4253–4260.

9. Gandra RF, Simão RC, Matsumoto FE, da Silva BC, Ruiz LS, da Silva EG, et al. Genotyping by RAPD-PCR analyses of Malassezia furfur strains from pityriasis versicolor and seborrhoeic dermatitis patients. Mycopathologia. 2006. 162:273–280.

10. Guillot J, Deville M, Berthelemy M, Provost F, Guého E. A single PCR-restriction endonuclease analysis for rapid identification of Malassezia species. Lett Appl Microbiol. 2000. 31:400–403.

11. Makimura K, Tamura Y, Kudo M, Uchida K, Saito H, Yamaguchi H. Species identification and strain typing of Malassezia species stock strains and clinical isolates based on the DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. J Med Microbiol. 2000. 49:29–35.

12. Sugita T, Kodama M, Saito M, Ito T, Kato Y, Tsuboi R, et al. Sequence diversity of the intergenic spacer region of the rRNA gene of Malassezia globosa colonizing the skin of patients with atopic dermatitis and healthy individuals. J Clin Microbiol. 2003. 41:3022–3027.

13. Jang SJ, Lim SH, Ko JH, Oh BH, Kim SM, Song YC, et al. The investigation on the distribution of Malassezia yeasts on the normal Korean skin by 26S rDNA PCR-RFLP. Ann Dermatol. 2009. 21:18–26.

14. Lee YW, Kim SM, Oh BH, Lim SH, Choe YB, Ahn KJ. Isolation of 19 strains of Malassezia dermatis from healthy human skin in Korea. J Dermatol. 2008. 35:772–777.

15. González A, Sierra R, Cárdenas ME, Grajales A, Restrepo S, Cepero de Garcia MC, et al. Physiological and molecular characterization of atypical isolates of Malassezia furfur. J Clin Microbiol. 2009. 47:48–53.

16. Zomorodian K, Mirhendi H, Tarazooie B, Zeraati H, Hallaji Z, Balighi K. Distribution of Malassezia species in patients with psoriasis and healthy individuals in Tehran, Iran. J Cutan Pathol. 2008. 35:1027–1031.

17. Mirhendi H, Makimura K, Zomorodian K, Yamada T, Sugita T, Yamaguchi H. A simple PCR-RFLP method for identification and differentiation of 11 Malassezia species. J Microbiol Methods. 2005. 61:281–284.

18. Clift DC, Dodd HJ, Kirby JD, Midgley G, Noble WC. Seborrheic dermatitis and malignancy. An investigation of the skin flora. Acta Derm Venereol. 1988. 68:48–52.
19. Lim SW, Shin MG, Lim JY, Yun SJ, Kim SJ, Lee SC, et al. Nested PCR for detection of Malassezia species from patient skin scales and clinical strains. Korean J Dermatol. 2008. 46:446–452.
20. Sugita T, Suzuki M, Goto S, Nishikawa A, Hiruma M, Yamazaki T, et al. Quantitative analysis of the cutaneous Malassezia microbiota in 770 healthy Japanese by age and gender using a real-time PCR assay. Med Mycol. 2010. 48:229–233.

21. Gaitanis G, Velegraki A, Alexopoulos EC, Chasapi V, Tsigonia A, Katsambas A. Distribution of Malassezia species in pityriasis versicolor and seborrhoeic dermatitis in Greece. Typing of the major pityriasis versicolor isolate M. globosa. Br J Dermatol. 2006. 154:854–859.





 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download