Abstract
Objectives
This study developed an integrated database for 15 regional biobanks that provides large quantities of high-quality bio-data to researchers to be used for the prevention of disease, for the development of personalized medicines, and in genetics studies.
Methods
We collected raw data, managed independently by 15 regional biobanks, for database modeling and analyzed and defined the metadata of the items. We also built a three-step (high, middle, and low) classification system for classifying the item concepts based on the metadata. To generate clear meanings of the items, clinical items were defined using the Systematized Nomenclature of Medicine Clinical Terms, and specimen items were defined using the Logical Observation Identifiers Names and Codes. To optimize database performance, we set up a multi-column index based on the classification system and the international standard code.
Results
As a result of subdividing 7,197,252 raw data items collected, we refined the metadata into 1,796 clinical items and 1,792 specimen items. The classification system consists of 15 high, 163 middle, and 3,588 low class items. International standard codes were linked to 69.9% of the clinical items and 71.7% of the specimen items. The database consists of 18 tables based on a table from MySQL Server 5.6. As a result of the performance evaluation, the multi-column index shortened query time by as much as nine times.
Conclusions
The database developed was based on an international standard terminology system, providing an infrastructure that can integrate the 7,197,252 raw data items managed by the 15 regional biobanks. In particular, it resolved the inevitable interoperability issues in the exchange of information among the biobanks, and provided a solution to the synonym problem, which arises when the same concept is expressed in a variety of ways.
Biomedical research requires correlations analysis between the effects of genes and diseases and the use of patient clinical data and specimen test results [12345]. Clinical studies increasingly rely on more advanced information technology for the analysis, exchange, and management of various biomedical data [6]. Around the world, there are established regional biobanks that have collected information on certain human diseases and large amounts of bio-data [7].
Biobanks use clinical items to build databases to handle quality inspection as well as other information and to store human samples and data obtained from donors with various conditions [8]. Biobanks that provide specimens and information play important roles in research for the prevention of diseases and in the development of new drugs and personalized therapy approaches [910].
Such biobanks cover a wide range, from broad population-based collections to specimen items for particular diseases. The ranges of diseases and specimen collections differ depending on the purpose of the institution (the main clinical service and research directions) [11]. That is, current biobanks are collecting various types of bio-data [712].
The data stored in biobanks are managed using Biobank Information Management Systems (BIMS). A BIMS is an object-oriented software architecture consisting of a multi-layered, large bio-dataset, including text and visual information [13]. It stores not only patient clinical information but also information relevant to human samples (sample handling and management information) [12].
However, cooperating biobanks cannot easily share information due to problems with the existing systems and fundamental problems with the databases [7], which often limit the scope and scale of collaborative studies performed [114]. This situation arises because most BIMS are inadequate in terms of exchanging information in a common format (syntactic interoperability) and comprehending information received from another system (semantic interoperability) [15].
These concepts have been highlighted by various practitioners and other institutions in addition to biobanks, but the biobanks in Korea are no exception. The Korean National Institute of Health (KNIH) organized a team of experts to envision the construction of biobanks in 2007, which has since supported the establishment of biobanks at university hospitals and has undertaken the Korean Biobank Project (KBP) to build the National Biobank of Korea (NBK) and network in 2008.
As a result, the KNIH was associated with the biobanks of 17 university hospitals, with NBK as the center [16]. KBP secured a total of 525,416 items of specimen information in December 2012. Of these, 325,952 were collected from the public through a cohort study; the other 199,464 represented patient information stored in the biobanks.
Each biobank can enter and manage human bio-data using the BIMS operated by the NBK. However, the BIMS manages the bio-data information by using a form limited to 18 items [17]. Since each biobank was designed to gather information independently for a prolonged period after its establishment, the biobanks share information with each other in spreadsheets. However, a spreadsheet is different from a database system. A researcher might make a simple mistake and not correct it. Therefore, over time, the uncertainty of the collected human resources information will inevitably increase. In addition, a spreadsheet cannot be accessed simultaneously by many researchers searching for bio-data information to match with various patient conditions.
The methodology of existing studies (managing bio-data information in a distributed environment with no common standard) cannot provide a large amount of high-quality bio-data, and researchers cannot ensure the interoperability of the bio-data information.
This study developed an integrated bio-data information database for analyzing the raw data managed by 15 biobanks in Korea to ensure syntactic and semantic interoperability. This study used 7,197,252 raw data items collected from 15 biobanks using three-step data analysis (specification, classification, and standardization) to define common standards and designed a database that can manage bio-data information. Specifically, three-step data analysis was used to design a data model that can purify and maintain the bio-data information gathered by analyzing large amounts of irregular data in biobanks.
Specification involves defining the metadata (data name, data type, value type, and specimen name, specimen type, tube, unit, reference value) by extracting items from the collected data. As metadata is structured data that are useful for database researchers, this data can be used to prevent typographical mistakes, increase understanding about a specific item, and search for bio-data information under various conditions.
Classification ensures syntactic interoperability to separate the detailed items into domains and concepts using a three-step classification (high, middle, and low). The classification system, as a common criterion suggested by us, can system-atically manage the biobank items.
Standardization guarantees semantic interoperability in conjunction with an international standard code appropriate to the area of each item. The linkage to an international standard code can clarify the meaning of the items and overcome the synonym problems faced by a global biobank. The synonym problem is the phenomenon in which the number of unnecessary item columns increases in a database when different biobanks describe the same item or concept in different ways.
Based on the metadata, classification system, and international standard code of an item, the integrated database manages and refines the irregular bio-data information. Specifically, the database is organized in a multi-column index based on the classification system and the international standard code in order to process the queries of large amounts of bio-data efficiently.
Unlike spreadsheets and BIMS, our proposed methodology differs from traditional research methodologies in that the high-quality bio-data information associated with an international standard code can be managed syntagmatically in a single database. In addition, the multi-column index can optimize database performance by connecting the interrelated columns based on the preferred term.
We held a briefing session to examine the holdings of the biodata collected from the 17 biobanks. The data collection was performed by describing the purposes, timelines, and methods of this study, with practitioners from each biobank in attendance. The method of data collection involved providing a USB drive to each biobank practitioner; these were delivered directly by mail with passwords to allow access only to researchers and practitioners after the data had been stored on the USB drives. The data collection was performed from April 2, 2014 to April 27, 2014 after approval was obtained from the Korea National Institute for Bioethics Policy.
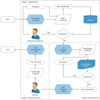
We selected this 'primitive' way, because there was the possibility of information leakage through hacking during the collection of large amounts of patient information via a web server. The types of data collected included questionnaires, case reports, collection methods, patient scenarios, and detailed items related to biodata to be managed from each biobank independently. Items of the collected data included data name, data type, value type, and specimen name, specimen type, specimen tube, units, reference values relevant to specimens, and additional explanations, which were the criteria that we used for the semantic definition of items to be used at the biobanks. The collected data were refined through three stages of specification, classification, and standardization for database modeling (Figure 1).
"Specification" was intended to define the metadata by extracting items from the collected data. For two or more of the same item, duplicate names were removed. Most items extracted were defined in metadata using the item specification; we also dealt with some omissions based on the data collected. For example, if the data type and value type of the items were missing, we analyzed data entered in the item and the pattern temporarily and received reviews from practitioners at each biobank. Defined metadata were validated by redistribution to each biobank.
We built a three-stage classification system based on the metadata to classify the items detailed as concept units (Figure 2). "High-class" is a top-level domain. High-class specimen items include diagnostic tests, functional tests, radiological tests, pathological examinations, and specimen information. High-class clinical items include epidemiological tests, physical information, social history, family history, past history, and present history. "Middle-class" is an intermediate concept included within specific high-class items, in which a classification name rule was applied to separate the subdivided concepts. For example, family history ('diseases') may be replaced with disease names, such as hepatoma, colon cancer, and stomach cancer, according to the item. "Low-class" refers to unique item names used in each biobank; it is the lowest concept of the classification system. To standardize the items that are represented by a unique abbreviation or code at each biobank, an international standard code for each field was associated with the item.
Three experts performed the linkage work using clinical items from the Systematized Nomenclature of Medicine Clinical Terms (SNOMED-CT), and two experts performed the linkage work using specimen items from the Logical Observation Identifiers Names and Codes (LOINC). The mapping work involved a process whereby SNOMED-CT was pre-coordinated mapped, and then post-coordinated mapped, cross-checked, and rechecked in order. LOINC involved a first mapping, second mapping, cross-check, and revalidation in order, based on the metadata and classification of the item.
Additionally, we introduced a solution to the synonym problem, in that items of the same concept were represented variously by each biobank, using associated international standard codes. Thus, we created the synonym grouped by data type and values of the item type, based on the same international standard code, and semantically minimized the number of redundant items by selecting representative terms.
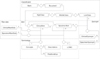
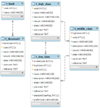
A researcher can refer to the classification system, the international standard code focusing on the database item, designed in conjunction with the raw data to allow access to the metadata of a specific item (Figure 3). The table is divided into five fields, depending on the role.
The 'Classification' field has five tables that store the document information, regional name of the biobank, and the classification of the items. Bank and Document have a relationship with 1:N. That is, all documents reflecting the uniqueness of each regional biobank are divided into distinct documents. HighClass, MiddleClass, and LowClass have relationships with each 1:N; if the upper concept is different, they are divided into different concepts even with the same classification name. In addition, Document and LowClass generate relationships of ClinicalItem and SpecimenItem to refer to the document and classification system information of a certain item.
The 'Item' field has two tables to store the metadata for an item. Each table includes the name of the item, the data type, and the value type, a description, the international standard codes, and a column for storing the classification information. Thus, at the center of all tables, ClinicalItem and SpecimenItem have relationships with various fields of the table to provide the metadata for an item in the particular raw data. Also, they have relationships with the table to store the raw data in the field RawData. The raw data table is composed of a large number of columns as well as the item table and primary key (PK) of a pair. This structure is designed for the purpose of refining data, which differs by the documents of each biobank, distinctive data types, value types of the same item, and errors in the entered data.
The 'Terminology' field has four tables to store the standard code of the international standard terminology system. SNOMED-CT has a connection between Relationships to store the relationships of concepts and Descriptions to store synonyms, preferred terms, and Fully Specified Name (FSN) in the center of Concepts. LOINC stores information about test names, units, times, samples, types, measures, and a Hangul translation, adding the full-name column and translation of the core columns. Concepts include ClinicalItem, and LOINC has SpecimenItem and relationships to provide more information about the item linked to the standard code.
The 'Synonym' field has two tables to store the synonym group and preferred term for an item. Each table stores items referring to the same code for two or more items associated with the standard code as synonyms.
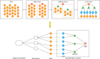
We designed a data model using three-step data analysis and database modeling. This solves the syntactic and semantic interoperability issues of bio-data information, the management of large amounts of bio-data, and the synonym problem noted in previous studies. The passage of an item of bio-data information through specification, classification, and standardization forms a three-dimensional information structure (Figure 4).
Each item consists of metadata, biobank, a formal document, classification systems, and an international standard code. The components are used in the detection and management of the stored bio-data information, in conjunction with the raw data table, and clarify the meaning of an item.
Metadata promotes better understanding of bio-data information, which is the criterion for refining the data. The name and description of metadata enable researchers to understand items and the test name, specimen type, tube, unit, and normal range ensure the reusability of the information.
The purpose of the classification system is to separate the items in the parent domain and the unit concept, which is the common standard for the biobanks in Korea classification system, which allows researchers to access the unit concept for an item and provides the basis for creating a new formal document in conjunction with metadata.
If a formal document is needed for a new research topic, researchers search for items to store the bio-data information through the classification and select the components of the formal document to be created by checking the metadata of the searched item. Since an item includes classification systems, as well as biobank and formal document information, it guarantees the syntactic interoperability of items transmitted during information exchange among the biobanks.
Since the international standard code defines the meanings of the items clearly, it allows the sharing of information with biobanks of another state. In addition, researchers can use the reference information (SNOMED-CT hierarchies and LOINC attributes) through the associated international standard code.
In this study, the classification system was built and mapped to standard terminology to ensure syntactic and semantic interoperability and to attempt to optimize the database performance. In the integrated database, it was difficult to apply theoretical methods (normalization and denormalization) because it was aggregated by various types of biobank columns and complex, heterogeneous clinical data. The multi-column index can optimize query time by grouping multiple columns into a single index. Therefore, it was important to set up the suitable index number for interrelated columns. This study set up the preferred term column to represent them as the index after grouping the synonym columns with the same standard code, and set up the index of the preferred terms based on the high-class concepts (Figure 5).
Data collection was accepted by 15 of the 17 biobanks; the remaining two rejected the data provided, because they were written by hand or were already managed bio-data using a database. In total, we collected 7,197,252 raw data items (146,403 patient cases and 104 documents). Considering the patient cases from the two biobanks that refused, we almost reached a total of 199,464 cases of the 17 biobanks, which were obtained by KBP in December 2012.
The 15 biobanks managed bio-data for all patients using spreadsheets, although the departments, researchers, and disease groups within the institutions used many spreadsheets in various types of documents. Documents were divided into 13 basic documents to collect basic personal demographics and essential items on the patients, 57 disease documents to separate the bio-data according to disease, 15 departmental documents to separate the bio-data according to the department that collected them, and 19 other documents collected individually by specific researchers.
Extracting items from the collected data resulted in a total of 3,588 items (1,796 clinical items and 1,792 specimen items). The classification system consisted of 15 high-class, 163 middle-class, and 3,588 low-class terms (Table 1). This classification system was structured based on entries from the 15 biobanks. Researchers can easily access items that are classified by concept, and the syntactic interoperability by the leveraging of information upon entry allowed exchanges among biobanks.
In the international standard code, 1,288 clinical items and 1,254 specimen items were linked to SNOMED-CT and LOINC, respectively (Table 1). The clinical items were composed of 1,170 pre-coordinated mappings and 118 post-coordinated mappings; the specimen items were composed of 1,222 single codes and 32 multiple codes. Types of non-mapping items were the question contents of the questionnaire, identification numbers, unsuitable concepts, and uncertain items. The question contents were unsuitable for the post-coordinated and multiple codes mappings because they were composed of a number of sentences. Identification numbers, such as local ID, BIMS ID, and box numbers, as well as unsuitable concepts, such as business names, could not be mapped to an international standard code. Uncertain items were concepts that can be mapped, but were suspended because of insufficient information (specimen type, tube, units, and reference values) collected. Uncertain items could not be mapped to a standard code exactly because LOINC has the different codes depending on the units, such as qualitative and quantitative, etc. This accounted for 5.2% of the total items. Considering the two cases of non-mapping, most items were mapped to the standard code.
An international standard code was associated with each item; based on the collected data, researchers and the system can clearly understand the concept of a specific item. In addition, 76 of the 826 synonyms in Clinical items and 127 of the 993 synonyms in Specimen items were selected, ultimately leaving only 203 preferred terms that could cover all of the 1,819 synonyms among the 15 biobanks.
In this study, we developed an integrated bio-data database using data analysis results, based on database modeling. The database developed using MySQL Server 5.6 comprises 18 tables. Depending on the relationships among the tables, they are divided into Classification, Clinical, and Specimen fields.
Unlike 'traditional' modeling, the database allows visualization of the PK of all taxonomy tables in t_clinical_item and t_specimen_item to store metadata for each item. Thus, the raw data are readily accessible.
The Classification field comprises five tables referenced in t_clinical_item and t_specimen_item (Figure 6). All tables have a code column PK to store sequential integers and a name column to store names. Additionally, the rest of the terms, except for t_bank, have a reference column for practitioners at the biobank to store notes and have the concept of state (add, delete, or change) in the comment column to indicate a change in status.
In the modeling of this study, t_low_class referred to the PK of t_high_class and t_middle_class to validate the upper and low classes together, unlike traditional modeling. The t_low_class has a column to store the PK of the selected preferred word in each synonym table, and Specimens have the flag column set at 1, whereas Clinical is 0, to separate the field of the item.
The Clinical field comprises seven tables in the center of t_clinical_item (Figure 7). The t_clinical_item refers to the PK of all classification system tables, and has data types for clinical items, value type, description, and a conceptId column.
Data types are divided into INT, DOUBLE, BOOLEAN, TEXT, DATETIME, and TIME, as used in MySQL, and value types symbolize the patterns of the raw data entered. For example, the value type of the sex item, entered as 'F' or 'M', is represented as 'selection (F, M)'. Because of this column configuration, a researcher can determine the classification and metadata by accessing items that make up the documents of a certain biobank. In addition, this architecture can ensure semantic interoperability by referring to synonyms from SNOMED-CT, the FSN, and layer information, using the code value of conceptId associated with the item.
In this study, we added t_clinical_item_refer to manage the meaning of an item and to manage a concept that includes two or more items as well as the meaning of an item that has been linked to uncertainty. The reason is that practitioners at the biobank are able to operate a post-coordinated operation regarding disconnected items with reference to the data of the html_formatted column.
The Specimen field comprises six tables in the center of t_specimen_item (Figure 8). The t_specimen_item also refers to the PK of the classification system table (t_high_class, t_middle_class, and t_low_class) and has a column to store the metadata of a specimen item. Unlike clinical items, we can see the detailed information on a specimen item from the column of t_loinc through the loinc_num column. In addition, PROPERTY, TIME_APSCT, SYSTEM, and SCALE_TYP columns store the Hangul information simultaneously by adding a FULLNAME and TRANSLATION column to each.
The t_specimen_raw_data stores the specimen item raw data in the biobank and many columns with a pair of values and item codes. For example, Urine_Protein_24hr is an item for which a value is input, and Urine_Protein_24hr_code is the PK of the t_specimen_item that is input. Thus, the system using the database developed can provide metadata and standard code information for the item, as well as raw data to a researcher.
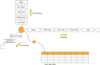
We tested the query speed and size of the database according to the multi-column index to evaluate the performance of the integrated database. The database is driven by a server computer that has an Intel Xeon E5420@2.5GHz (octa-core). The test method measured the single-query time and the size of the database according to the raw data. We confirmed that there as a significant difference in the query speed depending on whether or not the multi-column index was applied (Figure 9). The difference between the two cases could not be confirmed with 100,000 or fewer raw data items, since 500,000 raw data items was different from 2 times to 9 times.
These results were derived similarly from the figures assumed by us. This is because the ratio of preferred terms to synonyms was 1:9 (203 preferred terms:1,819 synonyms), we referred to this ratio to set up the 1 preferred column covering 9 synonym columns averagely. The database size was an average 2 times depending on whether or not the index was applied (Figure 10). The multi-column index can perform faster search because the index page has the data of the corresponding column.
However, if a column is wide, it is necessary to generate an index page for the number of the column, and this can result in waste of storage space as shown in Figure 10. Each time a data operation occurs, it is necessary to modify the index page as much as the size. That is, the multi column index is important to set up the index with an appropriate width; therefore, an analysis to generate the most appropriate index, such as classification and standardization, must be performed.
This study developed an integrated bio-database to provide high-quality biodata to researchers, by the specification, classification, and standardization of 7,197,252 raw data items from 15 biobanks. The database comprises entries separated via a classification concepts consisting of units, in conjunction with the international standard code for each item, and it stores large amounts of bio-data and metadata for the items to ensure interoperability. Thus, researchers will be able to more effectively contribute to biomedical advances by maximizing the utilization of bio-data information gathered in the biobanks.
Here, we were able to confirm the need for the construction of an infrastructure to ensure the interoperability of biodata, due to the problems found in research-phase processes. The data collected were mostly in the form of spreadsheets, and even practitioners within the same institution or the same biobank were managing patient bio-data in different ways. Unlike with such a database, the uncertainty of such information will increase over time, depending on what information they do or do not manage within Excel, because of its constrained capabilities. In particular, if the practitioner in charge or the supervisor changes, the uncertainty of the data may increase, because new rules and data management methods are used.
This phenomenon was confirmed by a case in which the deletion of certain items was requested because of the uncertainty of specific human resource information that was collected by practitioners at biobanks during the detailed verification of this study. The most efficient way to solve this problem is by using a database, rather than Excel. Thus, in this study, we developed an integrated database that can 'purify' the bio-data collected from each bank via an international standard terminology system.
The developed database was used to collect and analyze 146,403 cases from external researchers for the first time in comparison with patient information from 17 biobanks (199,464 cases) obtained from KBP in December 2012. Unlike previous studies, this study has great significance in that we used the international standard code to address interoperability issues that must be resolved in the exchange of information among biobanks. Also, keeping in mind the synonym problem in biobanks of other countries, we used the concept of SNOMED-CT and compressed the number of duplicate items semantically.
The system developed to date was designed to keep all data in one place without modification of the data kept in different types at each biobank. For further studies, it is important to normalize the raw data table as a concept unit, based on the preferred terms, designing the schema using Common Data Elements (CDE) and openEHR of Archetype of the National Cancer Institute. Also, extensive data maintenance with unanimous agreement among all biobanks should be performed to generally improve the accuracy of the data held.
Figures and Tables
Acknowledgments
This work was supported by the Research Program funded by the Korea National Institute for Bioethics Policy (Fund code: 2014-1).
References
1. Riegman PH, Morente MM, Betsou F, de Blasio P, Geary P. Marble Arch International Working Group on Biobanking for Biomedical Research. Biobanking for better healthcare. Mol Oncol. 2008; 2(3):213–222.

2. Jayasinghe SR, Mishra A, Van Daal A, Kwan E. Genetics and cardiovascular disease: design and development of a DNA biobank. Exp Clin Cardiol. 2009; 14(3):33–37.
3. Founti P, Topouzis F, van Koolwijk L, Traverso CE, Pfeiffer N, Viswanathan AC. Biobanks and the importance of detailed phenotyping: a case study: the European Glaucoma Society Glauco GENE project. Br J Ophthalmol. 2009; 93(5):577–581.

4. Voidonikolas G, Gingras MC, Hodges S, McGuire AL, Chen C, Gibbs RA, et al. Developing a tissue resource to characterize the genome of pancreatic cancer. World J Surg. 2009; 33(4):723–731.

5. Olund G, Lindqvist P, Litton JE. BIMS: an information management system for biobanking in the 21st century. IBM Syst J. 2007; 46(1):171–182.

6. Saltz J, Oster S, Hastings S, Langella S, Kurc T, Sanchez W, et al. caGrid: design and implementation of the core architecture of the cancer biomedical informatics grid. Bioinformatics. 2006; 22(15):1910–1916.

7. Hirtzlin I, Dubreuil C, Preaubert N, Duchier J, Jansen B, Simon J, et al. An empirical survey on biobanking of human genetic material and data in six EU countries. Eur J Hum Genet. 2003; 11(6):475–488.

8. Yuille M, van Ommen GJ, Brechot C, Cambon-Thomsen A, Dagher G, Landegren U, et al. Biobanking for Europe. Brief Bioinform. 2008; 9(1):14–24.

9. Zika E, Gurwitz D, Ibarreta D. Pharmacogenetics and pharmacogenomics: state-of-the-art and potential socio-economic impacts in the EU. Brussels, Belgium: Joint Research Center, European Commission;2006.
10. Austin MA, Harding S, McElroy C. Genebanks: a comparison of eight proposed international genetic databases. Community Genet. 2003; 6(1):37–45.

11. Spath MB, Grimson J. Applying the archetype approach to the database of a biobank information management system. Int J Med Inform. 2011; 80(3):205–226.

12. Watson PH, Wilson-McManus JE, Barnes RO, Giesz SC, Png A, Hegele RG, et al. Evolutionary concepts in biobanking - the BC BioLibrary. J Transl Med. 2009; 7:95.

13. Mora O, Bisbal J. BIMS: biomedical information management system [Internet]. [place unknown]: [publisher unknown];2013. cited at 2016 Mar 30. Available from: http://arxiv.org/abs/1303.5874.
14. Muilu J, Peltonen L, Litton JE. The federated database: a basis for biobank-based post-genome studies, integrating phenome and genome data from 600,000 twin pairs in Europe. Eur J Hum Genet. 2007; 15(7):718–723.

15. Mohanty SK, Mistry AT, Amin W, Parwani AV, Pople AK, Schmandt L, et al. The development and deployment of Common Data Elements for tissue banks for translational research in cancer: an emerging standard based approach for the Mesothelioma Virtual Tissue Bank. BMC Cancer. 2008; 8:91.




 PDF
PDF ePub
ePub Citation
Citation Print
Print







 XML Download
XML Download