I. Introduction
1. Hospital Information Systems and Electronic Medical Records
A hospital information system chiefly consists of an orderentry system, an administration system, a picture archiving and communication system (PACS), and electronic medical records (EMRs). Originally, Japanese hospital information systems consisted of just an electronic order-entry system and an administration system; the PACS and EMRs were added later. Most hospital information systems were developed by, and purchased from, computer companies. The prevalence of EMRs depends on the size of the hospital. The statistics for 2009 show that 825 major hospitals (with at least 400 beds) have the most advanced hospital information systems: 62.5% of their medical records are electronic [1]. Most new private clinics are equipped with EMR systems, especially in cities, yet EMRs make up only 16.5% of all their medical records. In medium-sized hospitals (100 to 399 beds), only 21.7% of medical records are electronic. In small hospitals (less than 99 beds), the EMR rate is just 9.1%.
Of the major hospitals, all 40 national university hospitals operate an EMR system but that is not the case for some of the hospitals belonging to private universities. The latter are hindered by financial restraints; however, from a hospital management point of view, they should be operating an EMR system because they are connected to other hospitals in regional networks. Few hospitals have achieved a complete paperless EMR system in the hospital because of the difficulty to construct in such as an intensive care unit or ophthalmology department. Major hospitals, however, intend to adopt a complete paperless EMR system.
2. Regional Networks
Japan's approach to EMR systems differs from other places such as Hong Kong. All the public hospitals in Hong Kong started electronic health records (EHRs) and EMRs at the same time. However, in Japan, various EMR systems were constructed by different companies, which mean the hospital information systems in Japan can only operate together through the connection of different platforms.
Several regional network systems have consequently been developed in Japan. The Dolphin system is one of the pioneering systems [2]: the data from each hospital are collected at the data center in the Medical Markup Language by means of Secure Socket Layer Virtual Private Network (SSL-VPN). The Superdolphin system provides a supersite that connects several Dolphin data centers so that doctors can see a patient's data in different geographical areas. Azisai-Net is one of the most successful regional network systems in Japan. It started as a one-way communication system to enable general practitioners or physicians in small hospitals to obtain the results of test data or images of special modality taken in a major hospital.
ID-LINK is a regional network that connects various facilities within a particular region. At present about 480 facilities are connected to the network. Every hospital opens its data in their server in demilitarized zone (DMZ); other hospitals can access the data via the data center.
Fujitsu's Humanbridge is a network system that connects hospitals via a data center on Security Architecture for Internet Protocol-Virtual Private Network (IPsec-VPN). Oshidori-Net is a system that shows the data of another hospital in the same display; it uses a thin client system to avoid the influence of other hospitals.
The Wakashio system has a minimum data set and is used specifically for patients with diabetes. PLANET is a system used in Kameda Hospital; it enables patients to access their EMRs.
Today there are several experimental systems with diverse styles and types, and there is no clear picture of the ultimate regional network. The regional networks constructed throughout Japan have mostly been design by industrial companies, which tend to regard the medicine and health field as the last available area for development. At present, however, the development of such networks is expensive, and complete interoperability offers few cost benefits for medical staff at the moment. Standardizing the laboratory data of various facilities is also a major problem. In addition, a regional network is similar to, but not completely the same as, an EHR system. And doctors recognize that complete interoperability is worthwhile long-term objective but not an initial requirement.
We have been constructing a system called IT net in Chiba University. It connects all the facilities of the Chiba prefecture for the exchange of referrals or images. IT net can connect only a limited amount of data, such as a simple text or image. Nevertheless, in view of its cost effectiveness for the medical profession, it might be a good solution for the time being.
3. EHRs, Personal Health Records, and the National Health Number
Regional healthcare information systems can provide more data than a single medical facility, and EHRs can be expanded to a national or global scale. The monthly data include the names of major diseases, the types and times of laboratory tests, and the names and doses of drugs administrated or injected in hospitals. The data can be collected electronically from all medical facilities in Japan and Korea. This information can be used to analyze national trends in the clinical treatment of particular diseases. EHRs can be used to electronically store all the events that affect a person's health throughout the course of the person's life. This type of record is called a personal health record (PHR) or a personal life record. A PHR includes a patient's entire health history: not only the medical data but also the health data.
Efforts have recently been made to start collecting the data of various facilities. Four national universities have now connected their laboratory data and diagnosis related group (DRG) data. The Pharmaceutical and Medical Devices Agency, Japanese version of the US Food and Drug Administration also plans to connect several major hospitals.
There are, however, several barriers to overcome in the creation of an EHR system. One major problem is the standardization of laboratory test data. Even in the same facility, many test measurements or units have changed over the last thirty years and they need to be calibrated for comparative purposes. Another problem is the right to individual privacy. Patient data must be subjected to a de-identification process to protect the confidentiality of the data. However, a set of data from several laboratories can be used to identify a patient even when personal data is removed. For the purpose of research and the establishment of a real national database, I believe the legislation needs to be amended to allow exceptional use of health records by facilities other than the patient's own facility. Unlike Korea, Japan does not use a social security number or a social health number. The Japanese cabinet has agreed to proceed with the introduction of a social security number but the issue is still controversial and there is no indication when it will be implemented.
The history of EMRs and EHRs in East Asian countries differs from that of Western countries. In East Asia, major hospitals began with an order-entry system, and that system gradually developed into an EMR system and later an EHR system. Western countries, on the other hand, especially northern European countries and the Netherlands [3,4], started with an EHR system or with a system that involved the electronic referral or transportation of images between clinics and hospitals. The hospitals themselves did not have an EMR system. The ultimate objective is the same but the East Asian approach and the Western approach followed different pathways to achieve the objective.
II. Text Mining of Discharge Summaries as a Reuse of EMR
Accumulated EMR data can be used for various studies and other purposes. Few studies to date have focused on EMR content, though there have been some trials. The term frequency-inverse document frequency (TF-IDF) method has been used for text mining of discharge summaries [5-7]. This method can help find similar cases in the literature or be used to check the adequacy of a diagnosis [8].
1. Text Mining of Discharge Summaries
For text mining of discharge summaries my colleagues and I began with a morphological analysis. Japanese sentences contain no spaces between words but the sentences can be divided into words with a Japanese morphological analysis tool such as MECAB [9]. Index terms and medical terms are then added to a special dictionary. The dictionary contains the medical dictionary and glossary of drugs, injections, and diseases is used at Chiba University Hospital.
Each word is weighted with the TF-IDF method, which is widely used in the field of information retrieval. In the TF-IDF method, document i and word j are expressed as follows:
where tf(ij) is the term frequency, idf(j) is the inverse document frequency, and N(i) is the document normalization coefficient.
The TF-IDF method expresses all case reports as vectors and forms a multidimensional vector called a vector space model (Figure 1). It then calculates the degree of similarity in documents defined as inner products between vectors (0 ≤ similarity degree ≤ 1).
2. Retrieval of Similar Case Reports from the Naikagakkai Archives
The retrieval of similar cases is one of the most important and beneficial processes for clinicians when they encounter a difficult case for diagnosis or treatment. However, most databases of case records are not accessible for comprehensive searches. The TF-IDF method was used in a morphological analysis of a database of more than 15,000 case reports extracted from the Japanese Society of Internal Medicine and MEDLINE. Japanese physicians can now use a new search tool for similar case retrieval based on text mining of the Web site of the Japanese Society of Internal Medicine. When the user first inserts the digitalized text of a case record into the dialog box, the text is morphologically analyzed and compared with each stored case on the basis of the calculated inner products. The relevant cases are sorted in terms of the degree of similarity. The user can obtain more detailed information and gain access to the authors of the similar case reports. In 2009, Japanese interns gained access to this system, which is called PINACO.
3. Matching of Diagnostic and Text Mining Results
Instead of searching similar case one by one from the data base, when comparing with the groups composed of many cases of same disease, the greatest similarity to the target summary is considered to be a diagnosis of the target case. An experiment confirmed that text mining of discharge summaries could be an effective means of making a diagnosis.
The diagnosis and procedure complex (DPC) is the Japanese DRG. The 14-digit number indicates the name of the disease and the type of treatment. The TF-IDF method is used to estimate the DPC codes from the discharge summaries. The correct diagnosis rate is calculated to divide the number of summaries of which DPC code estimated by TF-IDF is matched with the real DPC code by that of total summaries.
This experiment was based on the discharge summaries of three hospitals. The summaries of two hospitals (Chiba University Hospital and St. Luke's Hospital) were arranged according to the discharge dates and divided into two groups (text data group and test data group) at a ratio of 7:3; each group had at least 10 cases with the same DPC codes from both hospitals. The text data group was collected to generate a document vector space model based on the DPC; the test data group was collected to verify the automatic DPC selection. All the summaries from the third hospital, namely Saga University Hospital, were assigned to a second group. A total of 20,013 cases were used in this study. The cases contained 97 different DPC codes.
Correct diagnoses were made for more than 85% of the summaries from Chiba Hospital and St. Luke's Hospital. When the texts or model data were exchanged between the hospitals, the portion of correct diagnoses fell by approximately 10%. However, when a mixture of model data from both hospitals was used, the portion of correct diagnoses recovered to almost the same level as Chiba and St. Lukes' own model data. In the case of Saga University Hospital, where the model data are not based on that hospital's original summaries, the portion of correct diagnoses was much lower than that of the other two hospitals. However, when the mixture of model data from both hospitals was used, the portion of correct diagnoses was the same as the higher correct rate of two hospitals. Thus, the results confirm the text mining of summaries can be useful for automatic diagnosis in a screening process; they can also be used to build a universal model for every hospital.
4. Findings on Adverse Drug Reaction
One of the most anticipated uses of text mining is its ability to detect concealed complications or adverse reactions to drugs. The results of text mining were compared with changes in the laboratory data of patient with a real liver dysfunction. The first step was to collect all the terms in the summaries which conveyed the notion of liver dysfunction. From a total of 219,663 inpatients, we revealed that 4,721 (or 2.1%) inpatients had liver dysfunction during their admission from a laboratory database. To be precise, they were collected from laboratory data; explicitly the elevation of alanine aminotransferase more than 100 units while it was within normal range at the admission. Analysis of the discharge summaries by text mining led to the identification of liver dysfunction only 1,007 cases (0.45%) in 219,663 cases.
Similarly, the description of thrombocytopenia was detected in only 15.6% among the patients with platelets less than 30,000. Cases of diabetes (with HbA1c value greater than 6.0%) were identified in 57.5% from the description of discharge summaries. The results vary in relation to the diseases. Nevertheless, it is clear that in real conditions text mining is not highly effective for making correct diagnoses from the text of summaries. The quality of the discharge summaries is not as reliable as that of case reports.
Electronic summaries have the same low quality as paper discharge summaries [10-12]. However, the quality of electronic summaries is expected to improve in the near future as its importance is gradually recognized. Consequently, in spite of the current limitations of text mining, new and more effective text mining tools are expected to be available to clinical researchers within a few years.
III. Data-Driven and Knowledge-Driven Approach
The construction of clinical and public health databases of PHRs or EHRs has led to the accumulation of huge volumes of data, particularly in genetic research. Genetic research includes various types of analysis such as genome and sequence analysis and microarray data or genetic expression data analysis. And informatics plays a very important role in these types of analysis [13]. The rapidly emerging field of genetic research has spawned a huge amount of knowledge, which is stored in many genomic databases. Researchers use various techniques of informatics, such as data mining, to analyze the knowledge and discover new relations. Thus, data mining is an essential tool in this discipline.
Active multidisciplinary research is expected to boost biomedical informatics. In other words, the confluence of disciplines will lead to the discovery of new relations beyond the limits of individual disciplines. The complete DNA sequences represent one's intrinsic factors, while one's PHR includes one's extrinsic factors and final results. The ultimate objective is to connect these relations. The complete DNA sequences represent one's intrinsic factors, while one's PHR includes one's extrinsic factors and final results. The ultimate objective is to connect these relations [14]. Many steps and phases must be carried out to achieve this objective. Thus, specific tools must be developed to complete each step and each phase.
The traditional approach to biomedical science is a knowledge-driven approach. Hypotheses are generated from domain knowledge by coincidental experience or revolutionary inventions. In today's circumstances, however, there is a need for data-intensive science. Hypotheses can be generated automatically by applying computational science and inductive reasoning to enormous amounts of data [15]. These two approaches are not in conflict with each other. They can be combined or integrated to discover new knowledge (Figure 2). Thus, biomedical informaticians are expected to play a significant role in developing new methods in the field of data mining and machine learning and in making those methods available to domain experts.
Now Japan will have new super computer generation K series to assist data mining technique. By these techniques, after or even during the construction of PHR and EHR, biomedical informaticians would also act as supervisors and coordinators of biomedicine. Within the biomedical field, a new discipline must be developed for the purpose of comprehensively overseeing all the steps of biomedical informatics-from the micro level to the macro level of information. The new discipline must be used to identify which areas are unknown, which limiting factors remain to be solved, and which areas must be linked to other areas. These coordinators are not specific domain experts. They must fulfill their tasks by accelerating the progress of all biomedical science. As the current disciplines of biomedical informatics interconnect, new roles for biomedical and computer scientists will come to light.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


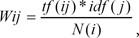

 XML Download
XML Download