Abstract
Calcineurin (CaN) is activated in diabetes and plays a role in glomerular hypertrophy and extracellular matrix (ECM) accumulation. Here, kidneys from diabetic model mice were investigated for the expression of the regulator of CaN 1 (RCAN1) isoform 4 (RCAN1.4) which had been shown to be transcriptionally upregulated by CaN activation. We found the increased immunoreactivity for RCAN1 in the glomerular cells of db/db mice and streptozotocin-induced diabetic mice. In concordance, the expression of RCAN1 protein and RCAN1.4 mRNA were elevated in the whole kidney sample from db/db mice. Interleukin-1β (IL-1β), tumor necrosis factor-α, and glycated albumin (AGE-BSA) were identified as inducers of RCAN1.4 in mesangial cells. Pretreatment of cyclosporine A blocked the increases of RCAN1.4 stimulated by IL-1β or AGE-BSA, suggesting that activation of CaN is required for the RCAN1.4 induction. Stable transfection of RCAN1.4 in Mes-13 mesangial cells upregulated several factors relevant to ECM production and degradation. These results suggested that RCAN1.4 might act as a link between CaN activation and ECM turnover in diabetic nephropathy.
References
1. Iidaka K, McCoy J, Kimmelstiel P. The glomerular mesangium: a quantitative analy-sis. Lab Invest. 1968; 19:573–579.
2. Kawano K, Arakawa M, McCoy J, Porch J, Kimmelstiel P. Quantitative study of glomeruli. Focal glomerulonephritis and diabetic glomerulosclerosis. Lab Invest. 1969; 21:269–275.
3. Ayo SH, Radnik RA, Garoni JA, Glass WF 2nd, Kreisberg JI. High glucose causes an increase in extracellular matrix proteins in cultured mesangial cells. Am J Pathol. 1990; 136:1339–1348.
4. Doi T, Vlassara H, Kirstein M, Yamada Y, Striker GE, Striker LJ. Receptor-specific increase in extracellular matrix production in mouse mesangial cells by advanced glycosylation end products is mediated via platelet-derived growth factor. Proc Natl Acad Sci USA. 1992; 89:2873–2877.

5. Mason RM, Wahab NA. Extracellular matrix metabolism in diabetic nephropathy. J Am Soc Nephrol. 2003; 14:1358–1373.

6. Flyvbjerg A. Putative pathophysiological role of growth factors and cytokines in experimental diabetic kidney disease. Diabetologia. 2000; 43:1205–1223.

7. Chiarelli F, Gaspari S, Marcovecchio ML. Role of growth factors in diabetic kidney disease. Horm Metab Res. 2009; 41:585–593.

8. Blázquez-Medela AM, López-Novoa JM, Martínez-Salgado C. Mechanisms involved in the genesis of diabetic nephropathy. Curr Diabetes Rev. 2010; 6:68–87.
9. Gr⊘nbaek H, Volmers P, Bj⊘rn SF, Osterby R, Orskov H, Flyvbjerg A. Effect of GH/IGF-I deficiency on long-term renal changes and urinary albumin excretion in diabetic dwarf rats. Am J Physiol. 1997; 272:E918–924.
10. Gooch JL, Barnes JL, Garcia S, Abboud HE. Calcineurin is activated in diabetes and is required for glomerular hypertrophy and ECM accumulation. Am J Physiol Renal Physiol. 2003; 284:F144–154.
11. Gooch JL, Tang Y, Ricono JM, Abboud HE. Insulin-like growth factor-I induces renal cell hypertrophy via a calcineurin-dependent mechanism. J Biol Chem. 2001; 276:42492–42500.

12. Gooch JL, Gorin Y, Zhang BX, Abboud HE. Involvement of calcineurin in transforming growth factor-beta-mediated regulation of extracellular matrix accumulation. J Biol Chem. 2004; 279:15561–15570.
13. Rothermel BA, Vega RB, Williams RS. The role of modulatory calcineurin-interacting proteins in calcineurin signaling. Trends Cardiovasc Med. 2003; 13:15–21.

14. Fuentes JJ, Genescà L, Kingsbury TJ, Cunningham KW, Pérez-Riba M, Estivill X, de la Luna S. DSCR1, overexpressed in Down syndrome, is an inhibitor of calcineurin-mediated signaling pathways. Hum Mol Genet. 2000; 9:1681–1690.

15. Fuentes JJ, Pritchard MA, Planas AM, Bosch A, Ferrer I, Estivill X. A new human gene from the Down syndrome critical region encodes a proline-rich protein highly expressed in fetal brain and heart. Hum Mol Genet. 1995; 4:1935–1944.

16. Fuentes JJ, Pritchard MA, Estivill X. Genomic organization, alternative splicing, and expression patterns of the DSCR1 (Down syndrome candidate region 1) gene. Genomics. 1997; 44:358–361.
17. Abe M, Sato Y. cDNA microarray analysis of the gene expression profile of VEGF-activated human umbilical vein endothelial cells. Angiogenesis. 2001; 4:289–298.
18. Mann KM, Ray JL, Moon ES, Sass KM, Benson MR. Calcineurin initiates smooth muscle differentiation in neural crest stem cells. J Cell Biol. 2004; 165:483–491.

19. Cho KO, Kim YS, Cho YJ, Kim SY. Upregulation of DSCR1 (RCAN1 or Adapt78) in the peri-infarct cortex after experimental stroke. Exp Neurol. 2008; 212:85–92.

20. Crawford DR, Leahy KP, Abramova N, Lan L, Wang Y, Davies KJ. Hamster adapt78 mRNA is a Down syndrome critical region homologue that is inducible by oxidative stress. Arch Biochem Biophys. 1997; 342:6–12.
21. Luo JD, Wang YY, Fu WL, Wu J, Chen AF. Gene therapy of endothelial nitric oxide synthase and manganese superoxide dismutase restores delayed wound healing in type 1 diabetic mice. Circulation. 2004; 110:2484–2493.

22. Bradshaw AD, Francki A, Motamed K, Howe C, Sage EH. Primary mesenchymal cells isolated from SPARC-null mice exhibit altered morphology and rates of proliferation. Mol Biol Cell. 1999; 10:1569–1579.

23. Kim YS, Cho KO, Lee HJ, Kim SY, Sato Y, Cho YJ. Down syndrome candidate region 1 increases the stability of the IkappaBalpha protein: implications for its anti-inflammatory effects. J Biol Chem. 2006; 281:39051–39061.
24. Kim YS, Lee HJ, Jang C, Kim HS, Cho YJ. Knockdown of RCAN1.4 increases susceptibility to FAS-mediated and DNA-damage-induced apoptosis by upregulation of p53 expression. Korean J Physiol Pharmacol. 2009; 13:483–489.

25. Hummel KP, Dickie MM, Coleman DL. Diabetes, a new mutation in the mouse. Science. 1966; 153:1127–1128.

26. Yang J, Rothermel B, Vega RB, Frey N, McKinsey TA, Olson EN, Bassel-Duby R, Williams RS. Independent signals control expression of the calcineurin inhibitory proteins MCIP1 and MCIP2 in striated muscles. Circ Res. 2000; 87:E61–68.

27. David KC, Scott RH, Nixon GF. Advanced glycation endproducts induce a proliferative response in vascular smooth muscle cells via altered calcium signaling. Biochem Pharmacol. 2008; 76:1110–1120.

28. Minami T, Horiuchi K, Miura M, Abid MR, Takabe W, Noguchi N, Kohro T, Ge X, Aburatani H, Hamakubo T, Kodama T, Aird WC. Vascular endothelial growth factor- and thrombin-induced termination factor, Down syndrome critical region-1, attenuates endothelial cell proliferation and angiogenesis. J Biol Chem. 2004; 279:50537–50554.

29. Simon M, Röckl W, Hornig C, Gröne EF, Theis H, Weich HA, Fuchs E, Yayon A, Gröne HJ. Receptors of vascular endothelial growth factor/vascular permeability factor (VEGF/VPF) in fetal and adult human kidney: localization and [125I]VEGF binding sites. J Am Soc Nephrol. 1998; 9:1032–1044.

30. Cooper ME, Vranes D, Youssef S, Stacker SA, Cox AJ, Rizkalla B, Casley DJ, Bach LA, Kelly DJ, Gilbert RE. Increased renal expression of vascular endothelial growth factor (VEGF) and its receptor VEGFR-2 in experimental diabetes. Diabetes. 1999; 48:2229–2239.

31. Wang P, Heitman J. The cyclophilins. Genome Biol. 2005; 6:226.
32. Mehta S, Li H, Hogan PG, Cunningham KW. Domain architecture of the regulators of calcineurin (RCANs) and identification of a divergent RCAN in yeast. Mol Cell Biol. 2009; 29:2777–2793.

33. Ke H, Huai Q. Structures of calcineurin and its complexes with immunophilins-immunosuppressants. Biochem Biophys Res Commun. 2003; 311:1095–1102.

34. Cho YJ, Abe M, Kim SY, Sato Y. Raf-1 is a binding partner of DSCR1. Arch Biochem Biophys. 2005; 439:121–128.

35. Lee EJ, Seo SR, Um JW, Park J, Oh Y, Chung KC. NF-kappaB-inducing kinase phosphorylates and blocks the degradation of Down syndrome candidate region 1. J Biol Chem. 2008; 283:3392–3400.
36. Seo SR, Chung KC. CREB activates proteasomal degradation of DSCR1/RCAN1. FEBS Lett. 2008; 582:1889–1893.

37. Asada S, Ikeda A, Nagao R, Hama H, Sudo T, Fukamizu A, Kasuya Y, Kishi T. Oxidative stress-induced ubiquitination of RCAN1 mediated by SCFbeta-TrCP ubiquitin ligase. Int J Mol Med. 2008; 22:95–104.
Fig. 1.
RCAN1.4 expression in the kidneys of diabetic mouse models. (A) Representative photomicrographs of RCAN1 immunohistochemistry in the glomerulus of kidney sections that were obtained from diabetic (db/db) and non-diabetic (db/m) mice at the age of 20 weeks, and from diabetic (STZ+) and non-diabetic (STZ-) C57BL mice at 16 weeks after treatment (400×). (B) Western blot analysis shows increased levels of RCAN1 protein in the whole kidney homogenates of diabetic (db/db) mice compared to non-diabetic (db/m) mice. Alpha-tubulin (α-tubulin) served as a loading control. (C) Specific induction of RCAN1.4 mRNA. The relative amount of mRNAs was estimated by real-time PCR, normalized by GAPDH, and the values for db/db mice (open bars) are relative to the expression observed in db/m mice (filled bars; arbitrarily designated a value of 1). Data are means±SE of five experiments. ∗p<0.01, db/m vs. db/db.
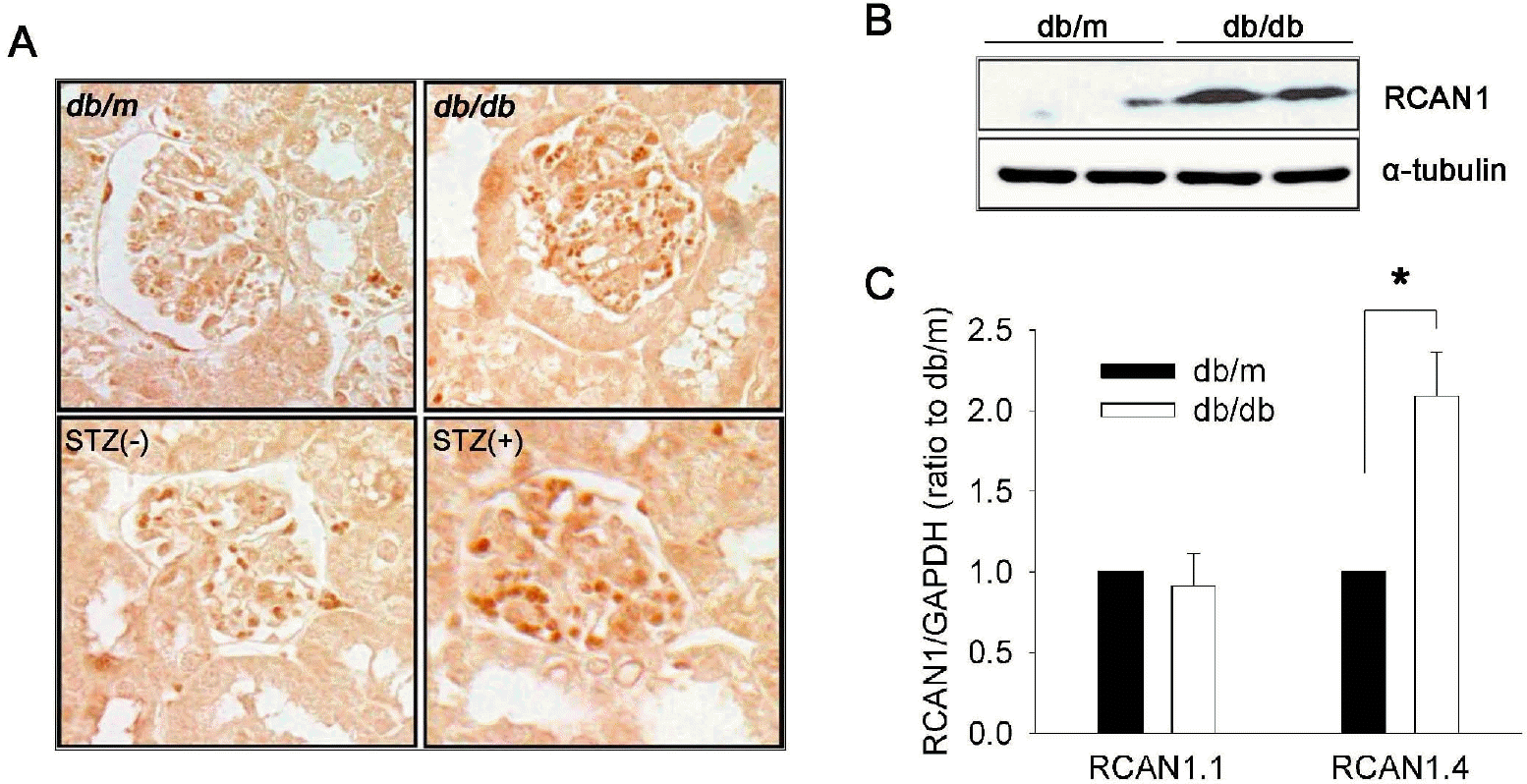
Fig. 2.
RCAN1.4 mRNA induction by diverse stimuli in mouse primary mesangial cells. (A) Mesangial cells were isolated from glomeruli by differential sieving, and then the cells of passages 7 to 10 in quiescent state were stimulated with the agents indicated below for 24 h or cultured for 48 h in high glucose condition (33 mM) or in osmotically balanced control medium (5.9 mM glucose plus 27 mM mannitol). Effect of the each stimulation was tested for RCAN1.4 mRNA induction, as described in the Methods. Concentrations were: TGF-β1 (4 ng/ml), ANG II (2μM), VEGF (10 ng/ml), IL-1β (20 ng/ml), TNF-α (20 ng/ml), MCP-1 (100 ng/ml), oxidative stress (300 μM of H2O2), BSA (50 μg/ml), and AGE-BSA (50 μg/ml). ∗p<0.01 compared to no-treatment. (B) Effect of CsA on IL-1β- or AGE-BSA-mediated RCAN1.4 induction was tested. Quiescent mesangial cells were stimulated with IL-1β (20 ng/ml) or AGE-BSA (50 μg/ml) in presence of CsA (1 μM; open bars) or vehicle (filled bars). After 24 h, mRNAs were isolated to determine RCAN1.4 mRNA induction. All the values are means±SE of five experiments/group. Values are expressed relative to the expression observed in untreated controls (arbitrarily assigned a value of 1). ∗p<0.01 vs. the corresponding control (vehicle) group.
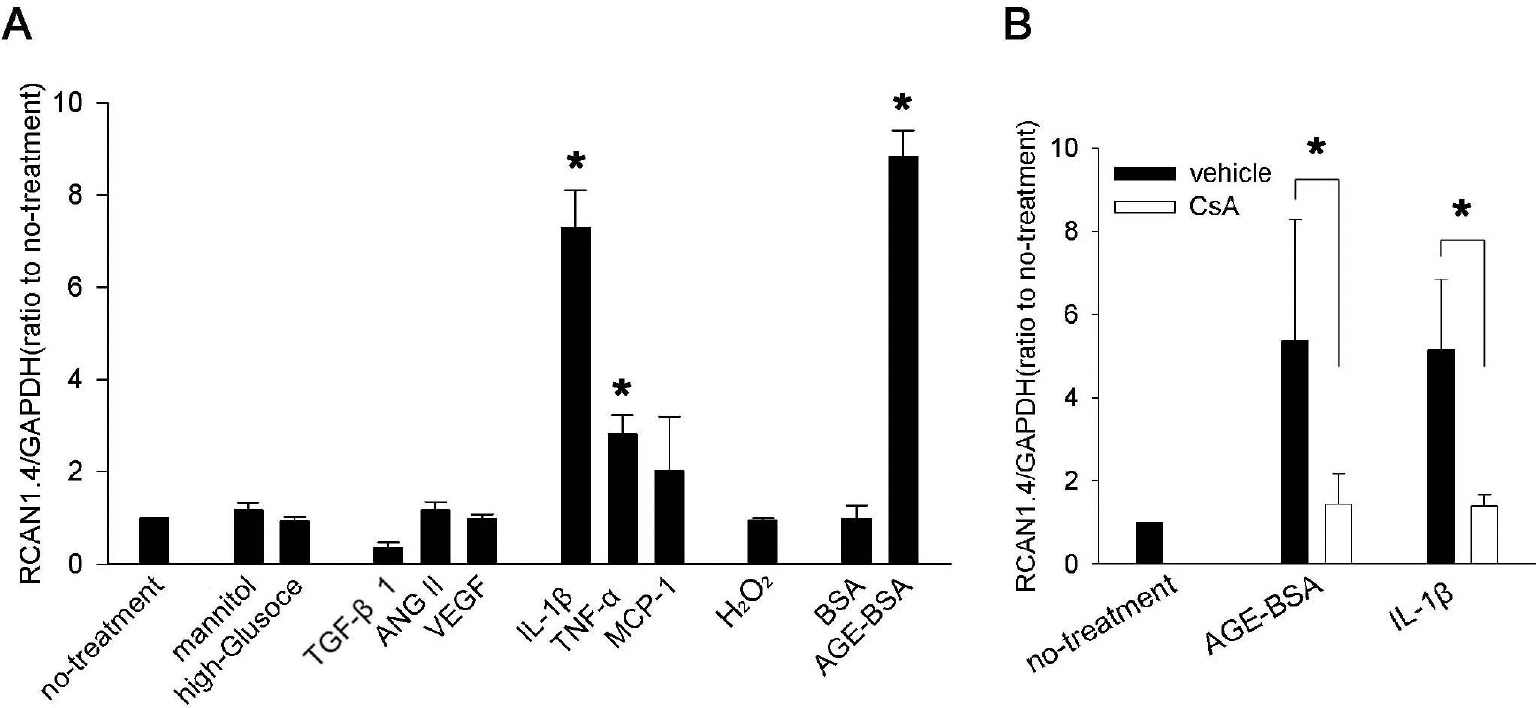
Fig. 3.
Expression patterns of ECM-related factors in Mes-13 cells stably transfected with RCAN1.4. Mes-13 cells were stably transfected with an expression vector containing RCAN1.4 construct (RCAN1.4; open bars) or vector alone (GFP; filled bars). From the actively dividing cells in complete medium, mRNAs were prepared and then subjected to analysis for the ECM-related gene expression with specific primers (Table 1). All values are means±SE of at least 5 experiments/group and the values of RCAN1.4-transfected cells are expressed relative to the expression observed in GFP-transfected cells (arbitrarily designated a value of 1). ∗p<0.01, RCAN1.4 vs. GFP group. Col, collagen.
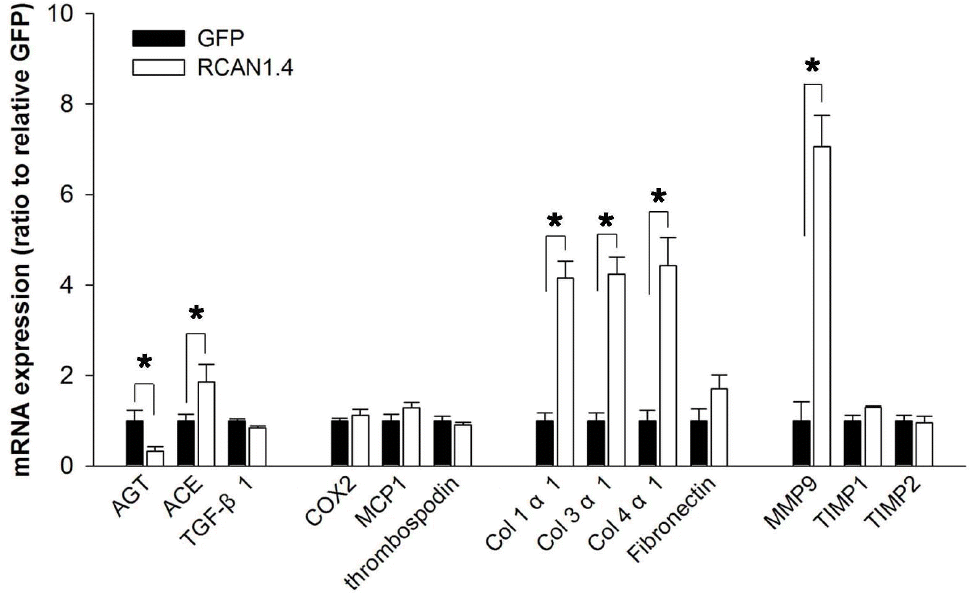
Table 1.
Primer sequences used for real-time RT PCR




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download