Abstract
The fyn-related kinase (FRK) belongs to the tyrosine kinase family of protein kinases. Recent studies have shown that Frk affects pancreatic beta cell number during embryogenesis and promotes beta cell cytotoxic signals in response to streptozotocin. To investigate the genetic association between FRK polymorphisms and the risk of obesity in Korean population, single nucleotide polymorphisms (SNPs) in the FRK gene region were selected and analyzed. The body mass index (BMI) was calculated, and biochemical data (systolic blood pressure, diastolic blood pressure, hemoglobin A1C, triglyceride, total cholesterol, high density lipoprotein, and low density lipoprotein) of blood sample from each subject were also measured. One hundred fifty five healthy control and 204 overweight/obesity subjects were recruited. Genotype frequencies of six SNPs [rs6568920 (+8391G>A), rs3756772 (+56780A> G), rs3798234 (+756870T), rs9384970 (+68506G>A), rs1933739 (+72978G>A), and rs9400883 (+ 75809A>G)] in the FRK gene were determined by Affymetrix Targeted Genotyping Chip data. According to the classification of Korean Society for the Study of Obesity, control (BMI 18 to < 23) and overweight/obesity (BMI ≥ 23) subjects were recruited. For the analysis of genetic data, EM algorithm, SNPStats, Haploview, HapAnalyzer, SNPAnalyzer, and Helixtree programs were used. Multiple logistic regression analysis (codominant, dominant, and recessive models) was performed. Age and gender as covariates were adjusted. For biochemical data, Student's t test was used. The mean value of BMI in the control and overweigh/obesity groups was 21.1±1.2 (mean±SD) and 25.6±2.0, respectively. All biochemical data of the overweight/obesity group were statistically significance, compared with the control group. Among six SNPs, two linkage disequilibrium (LD) blocks were discovered. One block consisted of rs1933739 and rs9400883, and the other comprised rs3756772 and rs3798234. One SNP (rs9384970, +68506G>A) showed an association with overweight/obesity in the codominant model (p=0.03). Interestingly, the AA genotype distribution in the overweight/obesity group (n=7, 3.5%) was higher than those in the control group (n=1, 0.6%), which is not found in either Japanese or Chinese subjects. Therefore, the AA genotype of rs9384970 may be a risk factor for development of obesity in Korean population. The results suggest that FRK may be associated with overweight/obesity in Korean population.
REFERENCES
Akerblom B., Anneren C., Welsh M. A role of FRK in regulation of embryonal pancreatic beta cell formation. Mol Cell Endocrinol. 270:73–78. 2007.
Alberti KG., Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 15:539–553. 1998.

Annerén C. Dual role of the tyrosine kinase GTK and the adaptor protein SHB in beta-cell growth: enhanced beta-cell replication after 60% pancreatectomy and increased sensitivity to streptozotocin. J Endocrinol. 172:145–153. 2002.
Annerén C., Welsh M. Increased cytokine-induced cytotoxicity of pancreatic islet cells from transgenic mice expressing the Src-like tyrosine kinase GTK. Mol Med. 7:301–310. 2001.

Annerén C., Welsh M., Jansson L. Glucose intolerance and reduced islet blood flow in transgenic mice expressing the FRK tyrosine kinase under the control of the rat insulin promoter. Am J Physiol Endocrinol Metab. 292:E1183–1190. 2007.

Barrett JC., Fry B., Maller J., Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 21:263–265. 2005.

Bell GI., Polonsky KS. Diabetes mellitus and genetically programmed defects in beta-cell function. Nature. 414:788–791. 2001.
Burton BT., Foster WR., Hirsch J., Van Itallie TB. Health implications of obesity: an NIH Consensus Development Conference. Int J Obes. 9:155–170. 1985.

Faul F., Erdfelder E., Lang AG., Buchner A. G∗Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 39:175–191. 2007.

Flegal KM., Carroll MD., Kuczmarski RJ., Johnson CL. Overweight and obesity in the United States: prevalence and trends, 1960-1994. Int J Obes Relat Metab Disord. 22:39–47. 1998.

Gabriel SB., Schaffner SF., Nguyen H., Moore JM., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., Liu-Cordero SN., Rotimi C., Adeyemo A., Cooper R., Ward R., Lander ES., Daly MJ., Altshuler D. The structure of haplotype blocks in the human genome. Science. 296:2225–2229. 2002.

Hubert HB., Feinleib M., McNamara PM., Castelli WP. Obesity as an independent risk factor for cardiovascular disease: a 26-year follow-up of participants in the Framingham Heart Study. Circulation. 67:968–977. 1983.

Jung HY., Park JS., Park YJ., Kim YJ., Kim K., Koh I. HapAnalyzer: minimum Haplotype analysis system for association studies. Genomics Inform. 2:107–109. 2004.
Kahn SE. The relative contributions of insulin resistance and beta-cell dysfunction to the pathophysiology of Type 2 diabetes. Diabetologia. 46:3–19. 2003.

Mokdad AH., Serdula MK., Dietz WH., Bowman BA., Marks JS., Koplan JP. The spread of the obesity epidemic in the United States, 1991-1998. JAMA. 282:1519–1522. 1999.

Nakano Y., Akahane A., Tanaka H., Ueno M., Kunugi H., Nanko S. Analysis of the fyn kinase gene in Alzheimer's disease and schizophrenia. No To Shinkei. 56:153–156. 2004.
Rybakowski JK., Borkowska A., Skibinska M., Hauser J. Polymorphisms of the Fyn kinase gene and a performance on the Wisconsin Card Sorting Test in schizophrenia. Psychiatr Genet. 17:201–204. 2007.

Solé X., Guinó E., Valls J., Iniesta R., Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 22:1928–1929. 2006.

Fig. 1.
Gene map and linkage disequilibrium (LD) in fyn-related kinase (FRK) gene. (A) Gene map of single nucleotide polymorphisms (SNPs) in FRK on chromosome 6q21-q22.3. Exons are marked with box. The coding regions are represented by black boxes and untranslation regions by white boxes. The first nucleotide of the transcriptional start site is denoted as +1. Asterisk (∗) indicates a significant SNP. Arrow indicates the location of each SNP. EX, exon. (B) LD coefficient (|D'|) and LD blocks among FRK SNPs. Block 1 consists of rs9400883 and rs1933739. Block 2 comprises rs3798234 and rs3756772.
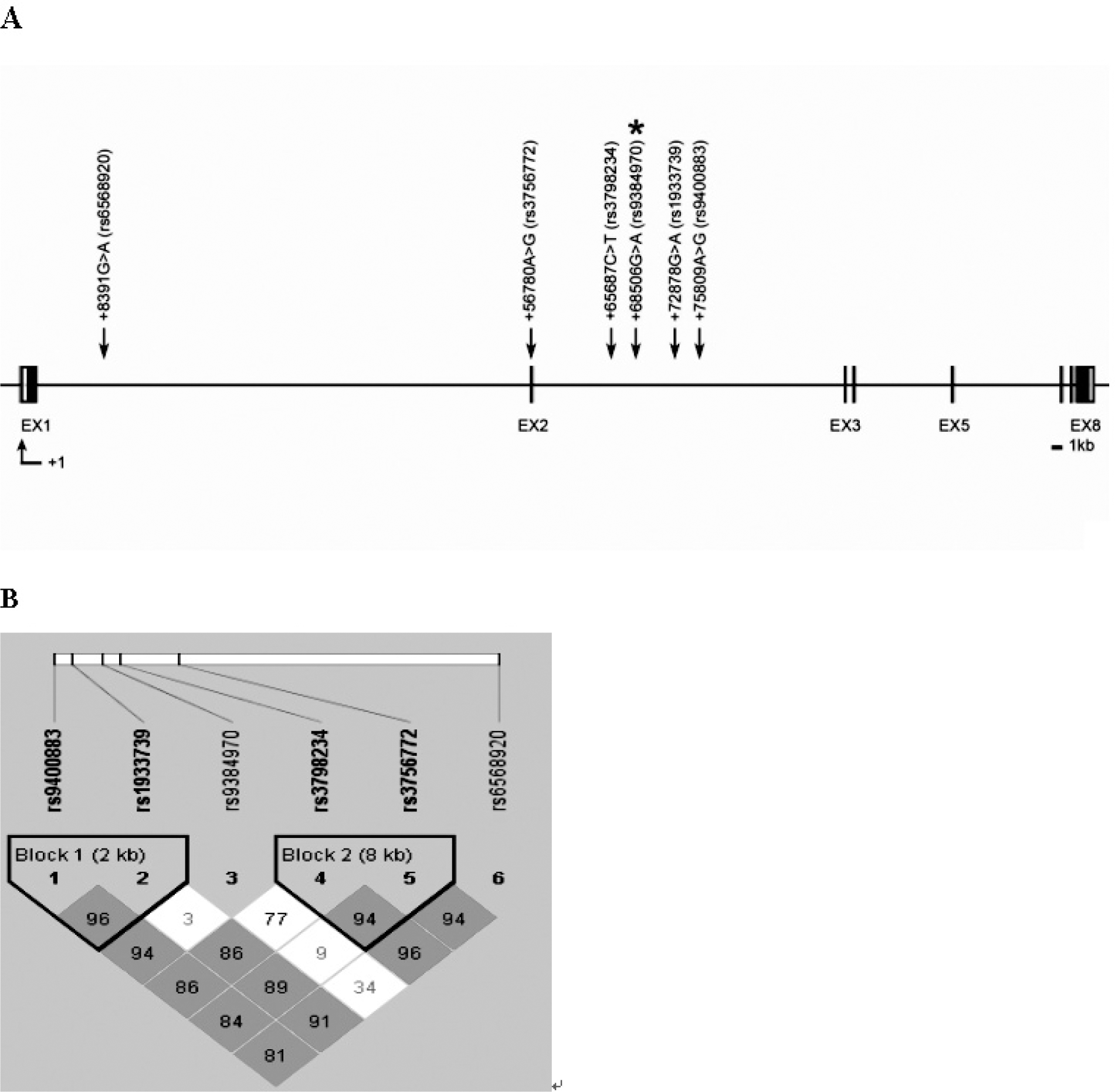
Table 1.
Clinical and biochemical characteristics of overweight/obesity and control subjects
Table 2.
Logistic regression analysis and genotype frequency of fyn-related kinase (FRK) polymorphisms in overweight/obesity and control subjects
Genotype distributions are shown as number (%). Odds ratio (OR), 95% confidence interval (CI), and p values were from logistic regression analysis with codominant, dominant, and recessive models controlling age and gender as covariates. Total number of each SNP is different, because genotypes of some SNPs are unreadable. SNP, single nucleotide polymorphism.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download