Abstract
REFERENCES
Figure 1.
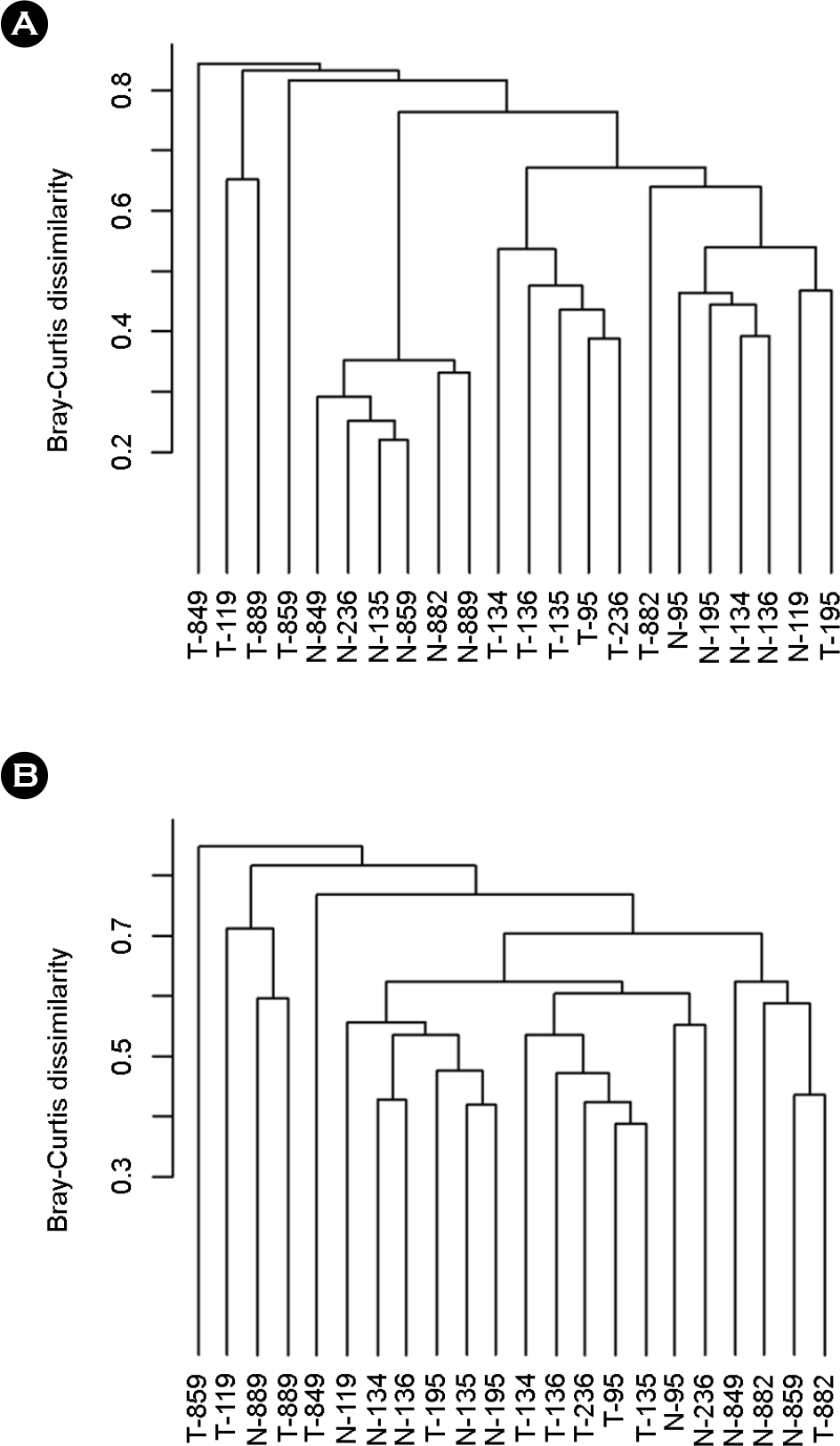
Figure 2.
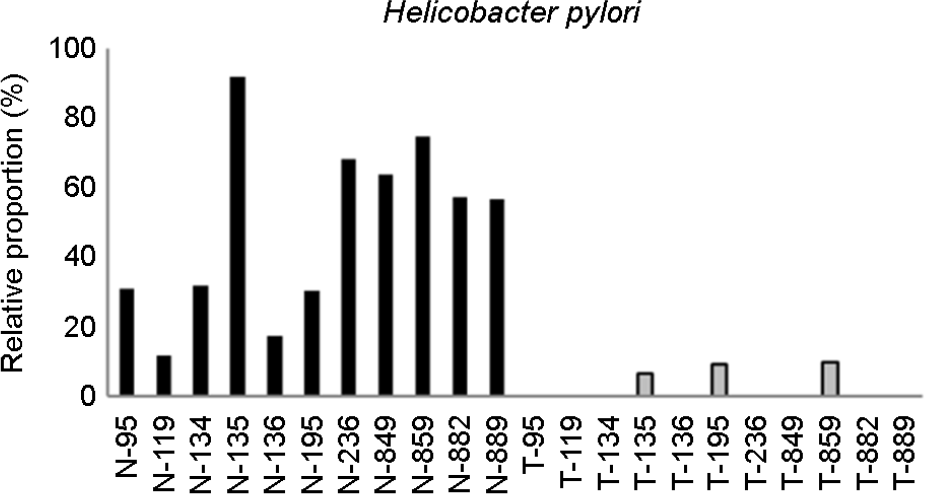
Figure 3.
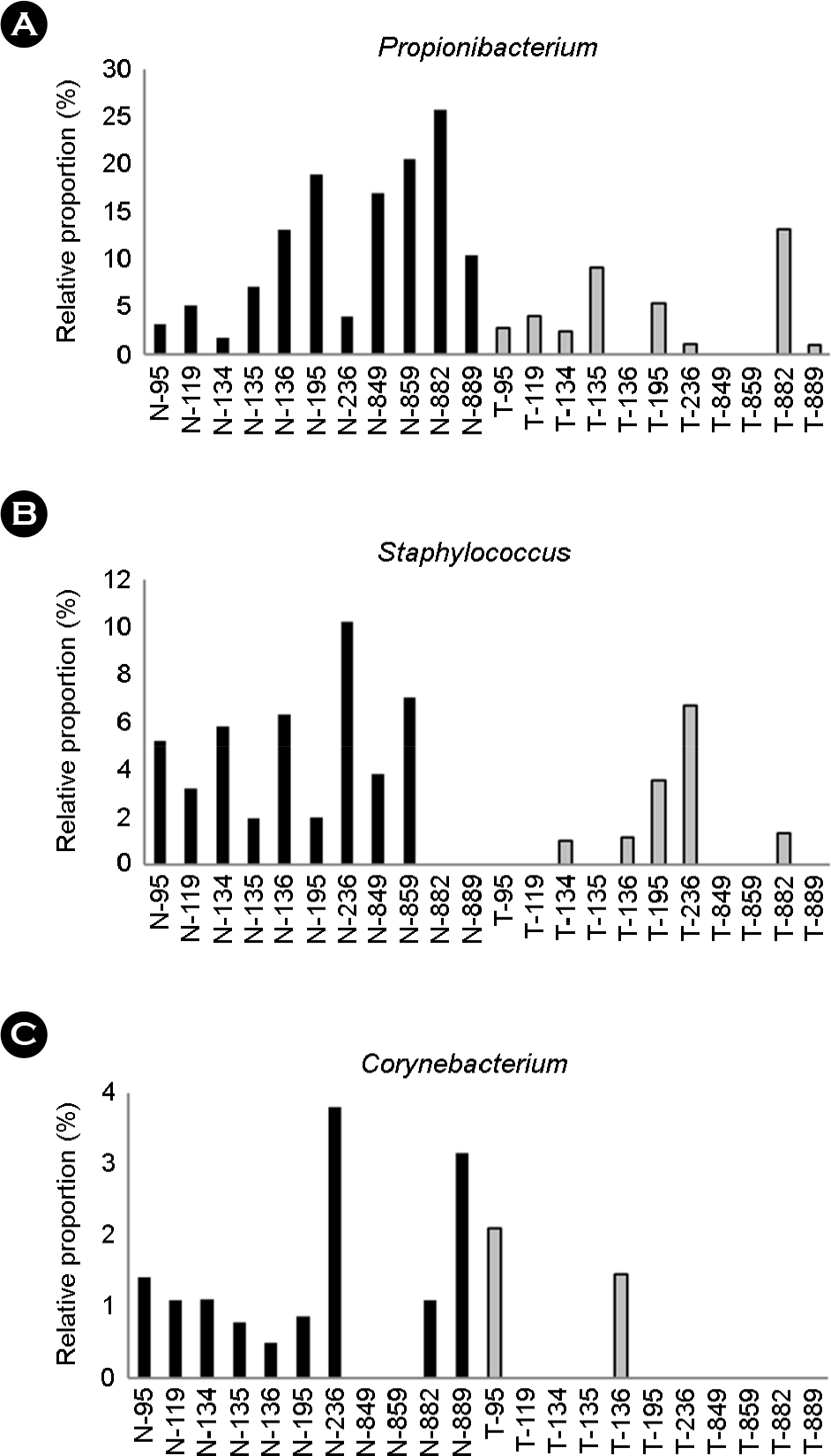
Figure 4.
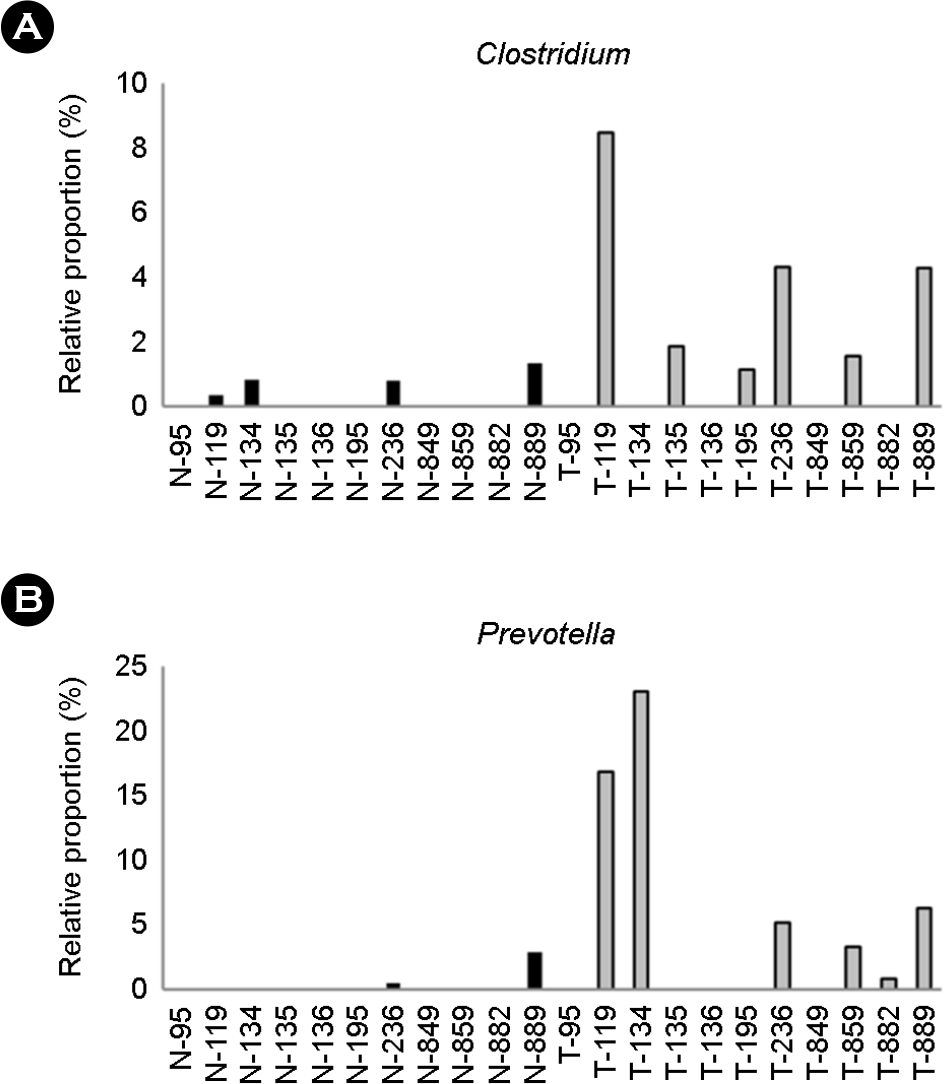
Table 1.
| Patient IDa | Sexa | Agea | Locationa | Size (cm)a | Histologic subtypea (WHO classification) | Laurena | Stage a | HP (%)b | Selectc |
|---|---|---|---|---|---|---|---|---|---|
| 43 | M | 51 | U | 7.0 | WD | Gastric | IIIb | 2.7 | |
| 80 | M | 56 | L | 5.0 | PD | Diffuse | IIIb | 1.3 | |
| 87 | M | 53 | U | 8.0 | MD | Diffuse | IIIb | 0 | |
| 95 | M | 65 | L | 6.0 | WD | Intestinal | IIIa | 31.1 | O |
| 119 | M | 48 | L | 10.0 | WD | Intestinal | IIIa | 11.7 | O |
| 130 | M | 57 | L | 6.5 | MD | Intestinal | IIb | 0 | |
| 134 | M | 57 | L | 8.5 | WD | Intestinal | IIIb | 31.8 | O |
| 135 | M | 37 | L | 6.0 | Mucinous | Intestinal | IIb | 92.0 | O |
| 136 | M | 55 | L | 4.5 | MD | Diffuse | IIIa | 17.5 | O |
| 195 | M | 67 | U | 6.0 | Papillary | Intestinal | IIIa | 30.4 | O |
| 236 | F | 72 | L | 6.5 | MD | Mixed | IIa | 68.1 | O |
| 849 | M | 64 | L | 5.4 | PD | Intestinal | Ib | 63.8 | O |
| 859 | F | 60 | L | 5.0 | MD | Intestinal | Ib | 74.8 | O |
| 882 | F | 75 | L | 3.5 | MD | Intestinal | Ia | 57.4 | O |
| 889 | F | 66 | L | 8.7 | MD | Intestinal | IIa | 56.6 | O |
| 917 | M | 73 | L | 7.9 | MD | Intestinal | IIa | 6.5 |
a Information marked were obtained from Yoon et al., Comprehensive genome- and transcriptome-wide analyses of mutations associated with microsatellite instability in Korean gastric cancers, Genome Res 23 (2013) 1109-1117. HP(%)
c Select, selected patients for analysis accroding to the presence of Helibacter pylori in the normal gastric mucosa; U, upper third; L, lower third; WD, tubular adenocarcinoma and well differentiated; MD, tubular adenocarcinoma and moderately differentiated; PD, tubular adenocarcinoma and poorly differentiated.
Table 2.
| SRA Runa | Patient IDb | Sampleb | Total readsc | Unmapped with hg19d | Unmapped read (%)e | Contigsf | Matching with bacteriag | Above thresholdh |
|---|---|---|---|---|---|---|---|---|
| SRR546237 | 43 | Normal | 52,088,890 | 13,775,741 | 26.4 | 98,994 | 1,722 | 246 |
| SRR546236 | 43 | Tumor | 52,088,890 | 12,407,493 | 23.8 | 33,734 | 388 | 49 |
| SRR825140 | 80 | Normal | 43,269,450 | 9,637,598 | 22.3 | 41,315 | 514 | 172 |
| SRR546238 | 80 | Tumor | 42,996,076 | 12,289,307 | 28.6 | 69,175 | 5,567 | 532 |
| SRR546239 | 87 | Normal | 44,471,834 | 14,136,479 | 31.8 | 93,583 | 1,227 | 190 |
| SRR546240 | 87 | Tumor | 49,814,912 | 12,612,740 | 25.3 | 70,767 | 3,171 | 227 |
| SRR825143 | 95 | Normal | 49,357,002 | 10,892,291 | 22.1 | 52,590 | 613 | 295 |
| SRR825141 | 95 | Tumor | 46,801,172 | 8,781,017 | 18.8 | 39,856 | 138 | 69 |
| SRR546226 | 119 | Normal | 52,088,890 | 14,958,865 | 28.7 | 62,186 | 2,739 | 327 |
| SRR546225 | 119 | Tumor | 55,022,224 | 16,085,525 | 29.2 | 35,455 | 135 | 58 |
| SRR825136 | 130 | Normal | 46,515,690 | 9,967,511 | 21.4 | 32,930 | 142 | 75 |
| SRR825135 | 130 | Tumor | 49,181,346 | 10,449,965 | 21.2 | 34,317 | 117 | 58 |
| SRR546227 | 134 | Normal | 52,400,004 | 12,290,105 | 23.5 | 31,281 | 500 | 97 |
| SRR546228 | 134 | Tumor | 55,022,224 | 13,180,350 | 24.0 | 34,657 | 252 | 91 |
| SRR546230 | 135 | Normal | 52,088,890 | 11,573,353 | 22.2 | 68,630 | 1,766 | 997 |
| SRR546229 | 135 | Tumor | 52,266,668 | 10,269,450 | 19.6 | 47,252 | 166 | 43 |
| SRR546232 | 136 | Normal | 54,755,556 | 14,774,615 | 27.0 | 66,951 | 845 | 177 |
| SRR546231 | 136 | Tumor | 53,511,112 | 10,454,368 | 19.5 | 26,586 | 166 | 66 |
| SRR546233 | 195 | Normal | 53,511,112 | 12,381,166 | 23.1 | 55,855 | 305 | 107 |
| SRR546234 | 195 | Tumor | 52,088,890 | 14,704,878 | 28.2 | 47,228 | 499 | 94 |
| SRR825139 | 236 | Normal | 41,204,902 | 9,277,569 | 22.5 | 44,692 | 857 | 498 |
| SRR825137 | 236 | Tumor | 43,956,792 | 6,135,987 | 14.0 | 20,651 | 192 | 89 |
| SRR801424 | 849 | Normal | 58,124,992 | 10,104,003 | 17.4 | 139,959 | 216 | 54 |
| SRR801425 | 849 | Tumor | 63,880,118 | 7,752,423 | 12.1 | 27,819 | 90 | 6 |
| SRR801427 | 859 | Normal | 59,019,350 | 10,010,433 | 17.0 | 114,625 | 356 | 104 |
| SRR801426 | 859 | Tumor | 56,227,424 | 6,982,330 | 12.4 | 86,906 | 188 | 57 |
| SRR801428 | 882 | Normal | 63,549,430 | 9,720,473 | 15.3 | 119,616 | 355 | 150 |
| SRR801429 | 882 | Tumor | 59,102,148 | 7,322,912 | 12.4 | 75,218 | 354 | 137 |
| SRR801430 | 889 | Normal | 44,498,444 | 7,450,469 | 16.7 | 88,745 | 412 | 122 |
| SRR801431 | 889 | Tumor | 59,655,378 | 7,622,162 | 12.8 | 25,708 | 881 | 134 |
| SRR801432 | 917 | Normal | 62,755,892 | 7,906,885 | 12.6 | 74,454 | 382 | 131 |
| SRR801433 | 917 | Tumor | 63,306,020 | 8,839,289 | 14.0 | 131,114 | 413 | 162 |
b Information marked with obtained from Yoon et al., Comprehensive genome- and transcriptome-wide analyses of mutations associated with microsatellite instability in Korean gastric cancers, Genome Res 23 (2013) 1109-1117.
Table 3.
| Genera | p-value |
|---|---|
| Helicobacter pylori | 0.000977 |
| Propionibacterium spp. | 0.013672 |
| Staphylococcus spp. | 0.014433 |
| Clostridium spp. | 0.034611 |
| Prevotella spp. | 0.036032 |
| Corynebacterium spp. | 0.044011 |
Significantly different microbiota in the population by comparing normal gastric mucosa and gastric cancer from the same patient. p-value for Helicobacter pylori was analyzed by comparing all strains at the species level. Other bacterial genera were analyzed at the genus level after removing all H. pylori sequences from the blast results. The Wilcoxon signed-rank test was applied for statistical analysis.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download