Abstract
To investigate the occurrence and distribution of serotype, specific virulence genes, and pulse field gel electrophoresis (PFGE) patterns in Vibrio parahaemolyticus isolates from Jeonnam, Korea, we tested 87 strains which were identified with V. parahaemolyticus from diarrheal episode patients in 2005. In this study, 16 different O:K serotype combinations of V. parahaemolyticus were determined. The distributions of O and K serotypes were O4:K68 (51.72%), O1:K70 (18.39%), O3:K6 (5.74%), O1:K68 (4.60%) and O3:K57 (4.60%) respectively. Serotype O4:K68 was the regional dominant specific serotype of V. parahaemolyticus in Sinan of Jeonnam, Korea. For the detection of thermostable direct hemolysin (tdh) and TDH-related hemolysin (trh) gene of V. parahaemolyticus, PCR was performed. The tdh gene was detected in all of the V. parahaemolyticus isolates from diarrheal patients, but trh gene was not detected. Analysis of PFGE patterns of 30 V. parahaemolyticus isolates showed 3 groups and 20 types. Among 14 O4:K68 serotypes which were isolated in Sinan, PFGE patterns of 12 strains were closely related (100%), but 2 strains were related by 58.3% and 45.4%, respectively. Also two strains of O1:K4 serotype in Gurye and two strains of O3:K6 serotype in Yeosu were closely related (100%), respectively. Although serotypes (O1:K4, O1:K70, O3:K6 and O4:K68) were different, PFGE patterns were related for more than 80.9%. Therefore, the epidemiological surveillance of V. parahaemolyticus is required by PFGE typing scheme as a further diagnostic tool.
REFERENCES
1). Su YC., Liu C. Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 2007. 24:549–58.
2). Honda T., Chearskul S., Takeda Y., Miwatani T. Immunological methods for detection of Kanagawa phenomenon of Vibrio parahaemolyticus. J Clin Microbiol. 1980. 11:600–3.
3). Takeda Y. Thermostable direct hemolysin of Vibrio parahaemolyticus. Pharmacol Ther. 1982. 19:123–46.
4). Kaysner CA., Tamplin ML., Twedt RM. Vibrio. Vanderzant C, Splittstoessr DF, editors. (Eds.),. Compendium of methods for the microbiological examination of foods. American Public Health Association;Washington, DC: 1992. p. 451–73.
5). Hondo S., Goto I., Minematsu I., Ikeda N., Asano N., Ishibashi M., Kinoshita Y., Nishhibuchi N., Honda T., Miwatani T. Gastroenteritis due to Kanagawa negative Vibrio parahaemolyticus. Lancet. 1987. 1:331–2.
6). Kishishita M., Matsuoka N., Kumagai K., Yamasaki S., Takeda Y., Nishibuchi M. Sequence variation in the thermostable direct hemolysin-related hemolysin (trh) gene of Vibrio parahaemolyticus. Appl Environ Microbiol. 1992. 58:2449–57.
7). Okuda J., Nishibuchi M. Manifestation of the Kanagawa phenomenon, the virulence-associated phenotype, of Vibrio parahaemolyticus depends on a particular single change in the promoter of the thermostable direct haemolysin gene. Mol Microbiol. 1998. 30:499–511.
8). Nishibuchi M., Taniguchi T., Misawa T., Khaeomaneelam V., Honda T., Miwatani T. Cloning and nucleotide sequence of the gene (trh) encoding the hemolysin related to the thermostable direct hemolysin of Vibrio parahaemolyticus. Infect Immun. 1989. 57:2691–7.
9). Honda T., Abad-Lapuebla MA., Ni YX., Yamamoto K., Miwatani T. Characterization of a new thermostable direct haemolysin produced by a Kanagawa-phenomenon-negative clinical isolate of Vibrio parahaemolyticus. J Gen Microbiol. 1991. 137:253–9.
10). Tada J., Ohashi T., Nishimura N., Shirasaki Y., Ozaki H., Fukushima S., Takano J., Nishibuchi M., Takeda Y. Detection of the thermostable direct hemolysin gene (tdh) and the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus by polymerase chain reaction. Mol Cell Probes. 1992. 6:477–87.
11). Wong HC., Lu KT., Pan TM., Lee CL., Shih DY. Subspecies typing of Vibrio parahaemolyticus by pulsed-field gel electrophoresis. J Clin Mocrobiol. 1996. 34:1535–9.
12). Chowdhury NR., Chakraborty S., Ramamurthy T., Nishibuchi M., Yamasaki S., Takeda Y., Nair GB. Molecular evidence of clonal Vibrio parahaemolyticus pandemic strains. Emerg Infect Dis. 2000. 6:631–6.
13). Tenover FC., Arbeit RD., Goering RV., Mickelsen PA., Murray BE., Persing DH., Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis criteria for bacterial strain typing. J Clin Microbiol. 1995. 33:2233–9.

14). Obata H., Kai A., Morozumi S. The trends of Vibrio parahaemolyticus foodborne outbreaks in Tokyo: 1989~2000. Kansenshogaku Zasshi. 2001. 75:485–9.
15). Bhuiyan NA., Ansaruzzaman M., Kamruzzaman M., Alam K., Chowdhury NR., Nishibuchi M., Faruque SM., Sack DA., Takeda Y., Nair GB. Prevalence of the pandemic genotype of Vibrio parahaemolyticus in Dhaka, Bangladesh, and significance of its distribution across different serotypes. J Clin Microbiol. 2002. 40:284–6.
16). Ju JW. Studies on Vibrio parahaemolyticus on the southern seas of Korea-on the isolation of V. parahaemolyticus from sea water, sea mud and marine products in Jeju, Keoje, Namhae, Yockji, Busan and Masan. J Korean Soc Microbiol. 1983. 18:1–9.
17). Ryu JK. Studies on biological characteristics of Vibrio parahaemolyticus isolated in Korea. Ph.D Thesis, Graduate School, Konkuk Univ. 1984.
18). Lee SJ., Chung JK., Lee DY., Lee BK., Lee KJ., Lee HM. A study of virulence factors and pathogenicity of Vibrio parahaemolyticus isolated from diarrheal patients and environmental sources in Gyeongbuk province. Infect Chemother. 2003. 35:407–15.
19). Nasu H., Iida T., Sugahara T., Yamaichi Y., Park KS., Yokoyama K., Makino K., Shinagawa H., Honda T. A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J Clin Micrbiol. 2000. 38:2156–61.
20). Kishishita M., Matsuoka N., Kumagai K., Yamasaki S., Takeda Y., Nishibuchi M. Sequence variation in the thermostable direct hemolysin-related hemolysin (trh) gene of Vibrio parahaemolyticus. Appl Environ Microbiol. 1992. 58:2449–57.
21). Kim YB. Virulence factors and genotyping of Vibrio parahaemolyticusis in sea water and clinical isolates. J Bacteriol Virol. 2001. 31:229–38.
22). Sakurai J., Matsuzaki A., Miwatani T. Purification and characterization of thermostable direct hemolysin of Vibrio parahaemolyticus. Infect Immun. 1973. 8:775–80.
23). Hara-Kudo Y., Sugiyama K., Nishibuchi M., Chowdhury A., Yatsuyanagi J., Ohtomo Y., Saito A., Nagano H., Nishina T., Nakagawa H., Konuma H., Miyahara M., Kumagai S. Prevalence of pandemic thermostable direct hemolysin-producing Vibrio parahaemolyticus O3:K6 in seafood and the coastal environment in Japan. Appl Environ Microbiol. 2003. 69:3883–91.
24). Food code. Korea Food and Drug Administration. 2005. ;518.
Figure 1.
PCR analysis of the tdh gene (198 bp) of the V. paraheamolyticus. M: molecular weight ladder (100 bp), lane 1: positive control (V. paraheamolyticus ATCC33816), lane 2: O1:K9, lane 3: O1:K13, lane 4: O1:K68, lane 5: O1:K70, lane 6:O2:K15, lane 7: O2:K19, lane 8: O3:K6, lane 9: O3:K57, lane 10:O4:K68, lane 11: O4:K68, lane 16: negative control.
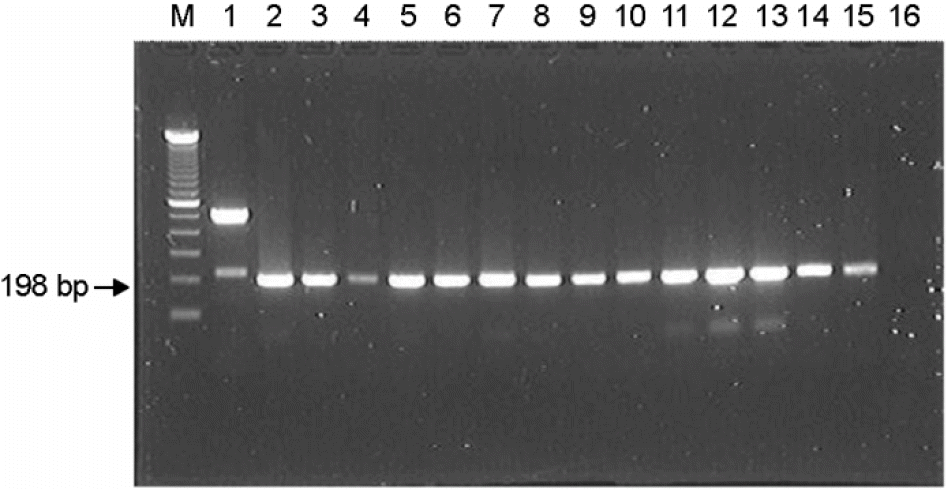
Figure 2.
PFGE patterns of enzyme (Not I) digested genomic DNA from V. parahaemolyticus isolates. M: Salmonella BAA-664, lane 1: O1:K4, lane 2: O1:K9, lane 3: O1:K13, lane 4: O1:K46, lane 5: O1:K56, lane 6: O1:K60, lane 7: O1:K61, lane 8: O1:K68, Lane 9: O1:K70, lane 10: O2:K15, lane 11: O2:K19, lane 12:O3:K6.
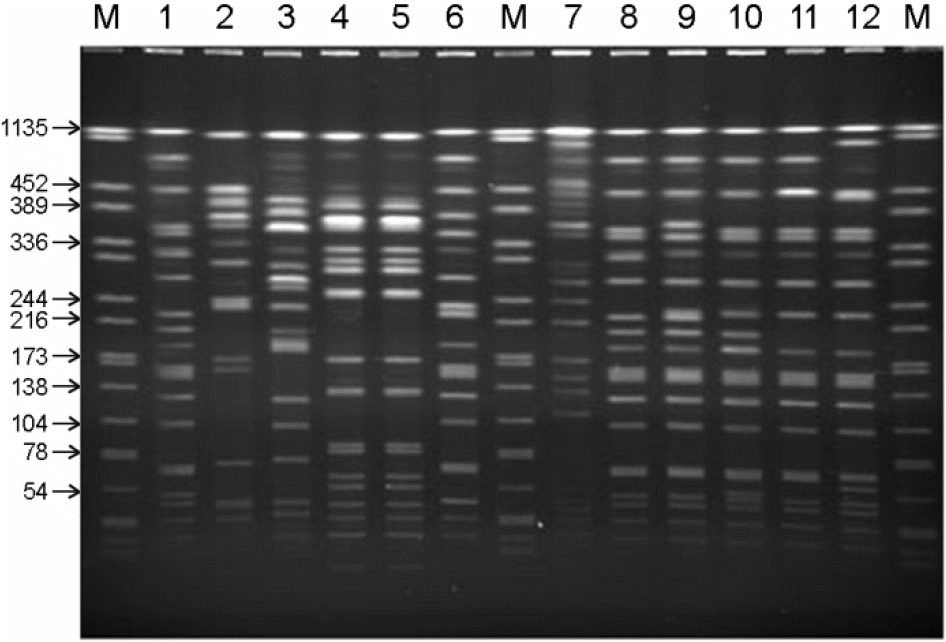
Figure 3.
Dendrogram of 30 strains of V. parahaemolyticus isolates as generated by UPGMA method from genetic distances.
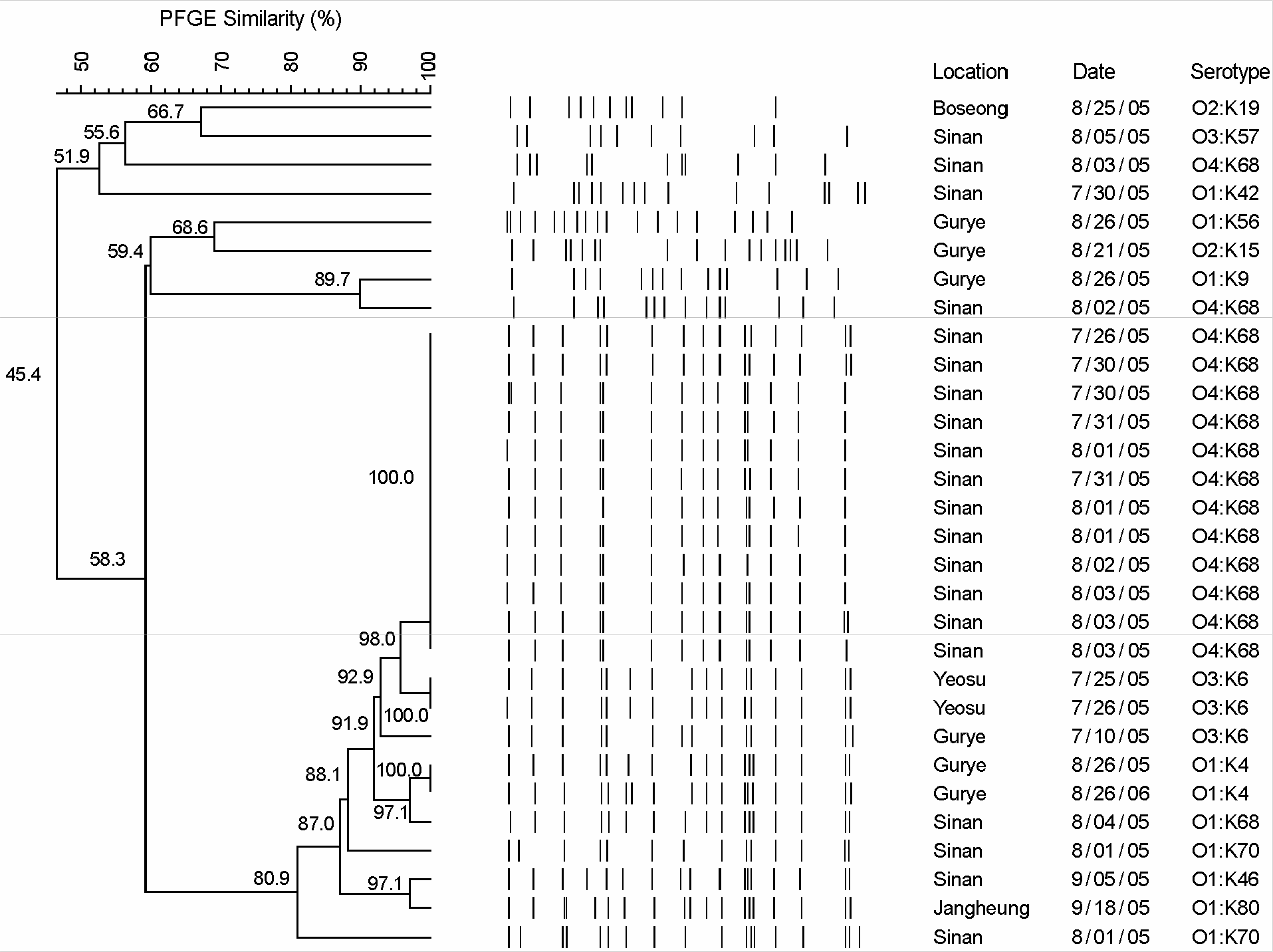
Table 1.
Synthetic oligonucleotides used in this experiment
| Target gene | 5′-3′ DNA sequence of primer | Amplicon size (bps) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| tdh | F | GGT | ACT | AAA | TGG | CTG | ACA | TCC | 198 |
| R | CCA | AAT | ACA | TTT | TAC | TTG | GAA | ||
| trh | F | GGC | TCA | AAA | TGG | TTA | AGC | GCC | 250 |
| R | CAT | TTC | CGC | TCT | CAT | ATG | CTT |
Table 2.
Occurrence and regional distribution of O and K serotype of V. parahaemolyticus isolated from diarrheal patients in Jeonnam, Korea
| Serotype | No. of strains | ||
|---|---|---|---|
| Total (%) | Sinan | Other regionsa | |
| O1:K4 | 1 (1.15) | 0 | 1 |
| O1:K9 | 3 (3.45) | 0 | 3 |
| O1:K42 | 1 (1.15) | 1 | 0 |
| O1:K46 | 1 (1.15) | 0 | 1 |
| O1:K56 | 1 (1.15) | 0 | 1 |
| O1:K60 | 1 (1.15) | 0 | 1 |
| O1:K61 | 1 (1.15) | 0 | 1 |
| O1:K68 | 4 (4.60) | 4 | 0 |
| O1:K70 | 16 (18.39) | 16 | 0 |
| O2:K15 | 1 (1.15) | 0 | 1 |
| O2:K19 | 1 (1.15) | 0 | 1 |
| O3:K6 | 5 (5.74) | 0 | 5 |
| O3:K8 | 1 (1.15) | 0 | 1 |
| O3:K57 | 4 (4.60) | 3 | 1 |
| O4:K29 | 1 (1.15) | 1 | 0 |
| O4:K68 | 45 (51.72) | 44 | 1 |
| Total | 87 | 69 | 18 |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download