Abstract
The present study describes the development of DNA vaccines using the hemagglutinin-neuraminidase (HN) and fusion (F) genes from AF2240 Newcastle disease virus strain, namely pIRES/HN, pIRES/F and pIRES-F/HN. Transient expression analysis of the constructs in Vero cells revealed the successful expression of gene inserts in vitro. Moreover, in vivo experiments showed that single vaccination with the constructed plasmid DNA (pDNA) followed by a boost with inactivated vaccine induced a significant difference in enzyme-linked immunosorbent assay antibody levels (p < 0.05) elicited by either pIRES/F, pIRES/F+ pIRES/HN or pIRES-F/HN at one week after the booster in specific pathogen free chickens when compared with the inactivated vaccine alone. Taken together, these results indicated that recombinant pDNA could be used to increase the efficacy of the inactivated vaccine immunization procedure.
Newcastle disease virus (NDV) is recognized as a highly infectious and fatal disease virus that affects many domestic and wild avian species [1]. Its entire genome consists of 15,186 nucleotides, with six structural genes in the order of 3'-NP-P-M-F-HN-L-5' that encode at least seven proteins [915]. The fusion (F) protein, which is the most immunogenic protein of the virus, plays important role in the beginning of infection by mediating fusion of the virus onto the host cell membrane and enabling viral entry into the cell membrane [1]. The hemagglutinin-neuraminidase (HN) protein is a multifunctional glycoprotein that plays important roles in virus attachment and fusion promotion activities [1]. The HN and F proteins produce virus neutralizing antibody responses and are the protective antigens [34]. The HN and F proteins are the main targets for immune response to NDV [13]. Vaccination programs for chickens usually consist of inactivated vaccines and/or live vaccines. Unfortunately, passive immunity is temporary and variable. Furthermore, vaccination with inactivated vaccines is time consuming, labor intensive, expensive, and often inaccurate.
DNA vaccination offers the same broad immunologic advantages as immunization with live, attenuated microorganisms, without the accompanying safety concerns. These vaccines have been shown to stimulate both humoral and cell-mediated immunity to the viral and bacterial pathogens that required both kinds of immune responses for protection [7]. Such vaccines have been studied for immunization against NDV strains. For example, Sakaguchi et al. [23] reported up to 40% protection against NDV using a single vaccination with the linearized NDV F gene; however, no protection was observed in response to the circular DNA plasmid expressing the F gene. In another study, Loke et al. [10] found high antibody titer with 50% protection after immunization with both linearized and circular plasmids expressing the F gene. Previous reports suggested that both F and HN glycoproteins interact with one another to promote fusion activity, and it is believed that immunization with a DNA plasmid encoding both proteins might induce an immune response to a broader spectrum of epitopes [25]. Therefore, the present study was conducted to investigate the induction of chicken immune response after immunization with three newly developed DNA vaccines encoding F, HN or both HN and F genes and boosted with inactivated NDV vaccine. The immunogenicity of these constructed DNA vaccines after one and two vaccinations was also investigated.
The AF2240 strain of NDV was propagated in the allantoic cavities of 10-day-old specific-pathogen-free (SPF) embryonated chicken eggs. Viral RNA was extracted from the allantoic fluid of the infected eggs 72 h post-inoculation using Trizol reagent. The purity and the concentration of the extracted RNA was determined based on the absorbance values at 260 and 280 nm.
The HN and F genes were amplified by reverse transcription polymerase chain reaction (RT-PCR) using the SuperScript III One-Step RT-PCR System with Platinum Taq High Fidelity kit (Invitrogen, USA) with the HN-specific primers 5'CAGTCGACGTCATGGGGAACCAGGCCTCACAA3' and 5'GAGCGGCCGCCCTATTGACAAGAATTCAGGCCAT3' and the F-specific primers 5'AATTCGGCTAGCACCATGG GCTCCAAGTCTT3' and 5'GGCACGCGTCTAGCTGCCA GAATTGACGCGCA3'. The primers were designed according to the published sequences of the genes (accession Nos. X79092 and AF048763, respectively). After reverse transcription at 45℃ for 45 min and an initial denaturation step at 94℃ for 3 min, PCR was conducted by subjecting the samples to 35 cycles of 30s at 94℃, 30s at 64℃ and 2 min at 72℃, followed by a final elongation step at 72℃ for 10 min.
The amplified fragments of the HN and F genes were inserted separately into the co-expression vector pIRES (Clontech, USA) to construct the DNA plasmids, pIRES-HN and pIRES-F, respectively. To construct the pIRES-F/HN plasmid, the HN and F genes were cloned into the SalI and NotI, and NheI and MluI sites of the vector, respectively. After transformation into Escherichia coli Top10 and selection on LB agar containing 50 µg/mL penicillin, the recombinant plasmids were extracted and subjected to double-stranded sequencing to verify the sequence and the correct orientation of the inserts. The constructs were purified using an endotoxin-free plasmid extraction kit and then resuspended in sterile endotoxin-free PBS.
The Vero cells were transfected with the constructs or the parental plasmid as a negative control using Lipofectamine LTX with Plus Reagent (Invitrogen) according to the manufacturer's instructions. The cells were tested for gene expression by Western blotting and indirect immunofluorescence.
Forty-eight hours post-transfection, the Vero cells were also tested by Western blot analysis using anti-NDV AF2240 polyclonal antibody raised in chicken as primary antibody and goat anti-chicken Ig Y conjugated to alkaline phosphatase (Abcam, USA) as secondary antibody.
The expression of the recombinant proteins was also studied by immunofluorescence tests as previously described [22], with some modifications [1112]. Briefly, the cells were washed with 1× phosphate-buffered-saline (PBS; pH 7.4) 48 h posttransfection, then fixed with 100% cold acetone for 10 min. Following three washes with 1× PBS, cells transfected with the pIRES/HN and pIRES/F plasmids were treated with chicken anti-NDV polyclonal antibody (Abcam), after which the cells transfected with the pIRES-F/HN plasmid were treated with anti-HN and anti-F monoclonal antibody for 1 h at RT. The cells were then washed again, after which they were overlaid with goat anti-chicken and a secondary fluorescein isothiocyanate (FITC)-labeled anti-chicken antibody (KPL, USA). Next, samples were washed twice with 1× PBS, then observed under an inverted fluorescence microscope.
Two-week-old SPF chickens were randomly divided into eleven different groups with 12 chickens in each group. Chickens were immunized via intramuscular injection of the plasmids into the pectoral muscle. The groups were subsequently vaccinated according to the following programs: group 1 with pIRES-F (50 µg pDNA); group 2 with pIRES-HN (50 µg pDNA); group 3 with pIRES-F/HN (100 µg pDNA); group 4 with pIRES-HN+pIRES-F (50 µg each pDNA); group 5 with pIRES-F (50 µg pDNA) + inactivated vaccine; group 6 with pIRES-HN (50 µg pDNA) + inactivated vaccine; group 7 with pIRES-F/HN (100 µg pDNA) + inactivated vaccine; group 8 with pIRES-F +pIRES-HN (50 µg each pDNA)+ inactivated vaccine; group 9 with pIRES (50 µg pDNA)+ inactivated vaccine; group 10 with inactivated vaccine alone and the last group was vaccinated with pIRES alone. All boosted groups were inoculated with 50 U of an inactivated NDV vaccine (Razi Institute, Iran) in Freund's Incomplete Adjuvant (FIA) at 30 days of age. Blood was collected from the wing vein of the chickens before and after vaccination for three weeks. Collected sera were stored at -20℃ for serological analysis.
DNA vaccines (pIRES-HN, pIRES-F and pIRES-F/HN) were constructed to express the HN and F genes of NDV strain AF2240 separately or synchronously. The constructs are shown in Fig. 1.
Western blot analysis confirmed the expression of the viral proteins in cells transfected with the recombinant plasmids. The HN protein (~74 kDa) was detected in cells transfected with pIRES-F/HN or pIRES-HN (Fig. 2). The NDV F protein was synthesized as a precursor, F0 protein. The glycosylated F0 protein migrates with an apparent molecular size of 64 KDa under nonreducing conditions or 66 under reducing conditions. The F protein must be cleaved to the mature F1+F2 form in order to function properly. However, the cleavage products, F1 polypeptide (55 kDa) and F2 polypeptide (12 kDa), remain linked by a disulfide bond [14].
An immunofluorescence test also showed the expression of the viral proteins. As shown in Fig. 3, cells transfected with the recombinant plasmids were observed by bright cytoplasmic green fluorescence, while no reaction was detected in the cells transfected with the parental plasmid.
ELISA antibody titer was evaluated at a serum dilution of 1 : 500 using an IDEXX ELISA kit (IDEXX Laboratories). The positive control mean was 0.165 and the negative control mean was 0.046. The S/P ratios calculated for each sample were as follow (Fig. 4):
All chickens vaccinated with pIRES/F/HN and pIRES/HN+ pIRES/F at 1, 2 and 3 weeks post DNA vaccination showed higher antibody responses than the control group (Fig. 4). At three weeks post vaccination, a significant increase (p < 0.05) in antibody response was detected in chickens vaccinated with the DNA plasmids and boosted two weeks later with the inactivated vaccine compared with non-boosted groups. The S/P ratio of ELISA titers in SPF chicken immunized with the inactivated vaccine alone was significantly lower (p < 0.05) than that of chickens immunized with pIRES/HN+pIRES/F and the pIRES/F/HN group boosted with inactivated vaccine at one week post booster.
Arora et al. [2] showed that immunization of chickens with NDV/HN+NDV/F viral proteins induced 73% protection against NDV challenge experiments, whereas 20% and 66% of chickens immunized with NDV/HN and NDV/F alone were protected against NDV challenge, respectively. However, 80% of chickens immunized with the whole virus vaccine survived on challenge [2]. Our findings are in line with those of earlier reports demonstrating that the presence of both HN and F antigen enable induction of the immune system better than HN or F alone [17].
Antibody titer in chickens immunized with pIRES/F plasmids at one week post-booster was higher (p < 0.05) than in chickens immunized with pIRES/HN (Fig. 4). These results may indicate greater immunogenicity of the NDV F gene than the NDV HN gene. The F gene is known to make a much greater contribution to NDV virulence than other genes [6].
Conversely, the higher antibodies titer developed in response to pIRES/F (the F gene inserted to site A of pIRES) than pIRES/HN (the HN gene inserted site to B of pIRES) may be related to the efficacy of pIRES plasmid. There is a cytomegalovirus promoter in pIRES plasmid that induced nuclear transcription from capped bicistronic mRNA which made with pIRES plasmid. In the present study the HN gene was inserted to B segment of multiple cloning site. In the B cistron, translation is down to the more modest level of the A cistron [19]. The results of the present study showed a low HN antibody titer than F antibody titer which was inserted to A cistron of pIRES. This low level of antibody can be considered in low translation gene due to down translation of B cistron site.
The highest Ab titers were detected in chickens vaccinated with pIRES/F/HN and then boosted with the inactivated vaccine. These results indicate that co-expression of HN and the F gene can enhance humoral responses to the virus and therefore be used to improve the efficiency of the inactivated vaccine. It is believed that F and HN glycoproteins interact with one another to promote fusion activity [25]; accordingly, immunization with the DNA plasmid co-expressing the genes or co-administration of both of the plasmid constructs expressing the F and HN proteins would be more effective than either component alone. Previous studies have also shown that co-expression of the both F and HN genes more effectively induces immune response to NDV [510]. However, the result of a previous study indicated that a bivalent pIRES plasmid containing both HN and F genes of NDV induced lower IFN-γ and cell-mediated immunity (CMI) responses than constructs containing single genes in maternal antibody-positive commercial chickens [18]. Both CMI and humoral response are thought to play a primary role against NDV infection in chickens [2021], although a recent study indicated that protection efficacy of a NDV DNA vaccine is more closely related to humoral immune response than CMI [5].
The efficacy of the DNA vaccines may be improved if they are applied together with plasmids expressing cytokines and/or costimulatory molecules [524]. Other approaches such as microencapsulation or association of the DNA plasmids nanoparticles can also be used to improve gene delivery and gene transfection in vivo, resulting in higher immune response [816].
In conclusion, the results of the present study also indicate that co-expressing the HN and F gene of NDV can induce immune response to the virus and improve efficiency of the available inactivated vaccine. The findings presented herein will provide further information regarding the potential value of DNA vaccines to NDV infection.
Figures and Tables
Fig. 1
Map of the DNA vaccines. The DNA plasmids were constructed by cloning the fusion (F) gene into the NheI and MluI\I sites and the hemagglutinin-neuraminidase (HN) gene into SalI and NotI of the expression vector pIRES.

Fig. 2
Western blot analysis. Vero cells were transfected with the constructs; 48 h post-transfection, gene expression was evaluated by Western blot analysis. (A) M, protein ladder; Lines 1–2, immunoblotting for the cells transfected with pIRES-F/HN, co-transfected with the DNA plasmids pIRES-HN and pIRES-F; Line 3, cells transfected with pIRES-F; Line 4, control cells transfected with the parental plasmid. (B) M, protein ladder; 1, immunoblotting of the cells transfected with virus as a positive control; Lines 2–3, cells transfected with pIRES-HN; Line 4, control cells transfected with the parental plasmid.

Fig. 3
Indirect immunofluorescence test. At 48 h post-transfection, vero cells transfected with pIRES-HN (B), pIRES-F (C), pIRES-F/HN (D) and/or the empty plasmid (A) as a negative control. Cells transfected with pIRES/HN and pIRES/F were treated with chicken anti-NDV polyclonal antibody, while cells transfected with pIRES-F/HN were treated with anti-HN and anti-F monoclonal antibody as the primary antibodies. Acll cells were then treated with FITC-conjugated goat anti chicken IgY as a secondary antibody. The cells were observed under the 20× objective of an inverted fluorescence microscope.
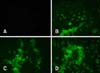
Fig. 4
S/P ratio column chart with SD bars per week as determined by ELISA. The error bars show the standard deviation of the means. Group 1, pIRES-F; Group 2, pIRES-HN; Group 3, pIRES-HN/F; Group 4, pIRES-HN+pIRES-F; Group 5, pIRES-F+inactivated vaccine; Group 6, pIRES-HN+inactivated vaccine; Group 7, pIRES-F/HN+inactivated vaccine; Group 8, pIRES-HN+pIRES-F+inactivated vaccine; Group 9, pIRES+ inactivated vaccine; Group 10, inactivated vaccine; Group 11, pIRES.

Acknowledgments
This work was funded by research grant ERGS/1-2012/5527122 and the Institute of Bioscience, Higher Institution Centre of Excellence (IBS HICoE) grant from the Ministry of Higher Education, Government of Malaysia.
References
1. Alexander D. Newcastle disease and other avian Paramyxoviridae infection. In : Calnek BW, Barnes HJ, Beard CW, McDougald LR, Saif YM, editors. Diseases of Poultry. 10th ed. Ames: Iowa State University Press;1997. p. 541–549.
2. Arora P, Lakhchaura BD, Garg SK. Evaluaion of immunogenic potential of 75kDa and 56kDa proteins of Newcastle disease virus (NDV). Indian J Exp Biol. 2010; 48:889–895.
3. Boursnell ME, Green PF, Samson AC, Campbell JI, Deuter A, Peters RW, Millar NS, Emmerson PT, Binns MM. A recombinant fowlpox virus expressing the hemagglutinin-neuraminidase gene of Newcastle disease virus (NDV) protects chickens against challenge by NDV. Virology. 1990; 178:297–300.

4. Cosset FL, Bouquet JF, Drynda A, Chebloune Y, Rey-Senelonge A, Kohen G, Nigon VM, Desmettre P, Verdier G. Newcastle disease virus (NDV) vaccine based on immunization with avian cells expressing the NDV hemagglutinin-neuraminidase glycoprotein. Virology. 1991; 185:862–866.

5. Donnelly JJ, Wahren B, Liu MA. DNA vaccines: progress and challenges. J Immunol. 2005; 175:633–639.

6. Dortmans JCFM, Koch G, Rottier PJM, Peeters BPH. Virulence of Newcastle disease virus: what is known so far? Vet Res. 2011; 42:122.

7. Gurunathan S, Klinman DM, Seder RA. DNA vaccines: immunology, application, and optimization. Annu Rev Immunol. 2000; 18:927–974.

8. Jazayeri SD, Ideris A, Zakaria Z, Shameli K, Moeini H, Omar AR. Cytotoxicity and immunological responses following oral vaccination of nanoencapsulated avian influenza virus H5 DNA vaccine with green synthesis silver nanoparticles. J Control Release. 2012; 161:116–123.

9. Krishnamurthy S, Huang Z, Samal SK. Recovery of a virulent strain of Newcastle disease virus from cloned cDNA: expression of a foreign gene results in growth retardation and attenuation. Virology. 2000; 278:168–182.

10. Loke CF, Omar AR, Raha AR, Yusoff K. Improved protection from velogenic Newcastle disease virus challenge following multiple immunizations with plasmid DNA encoding for F and HN genes. Vet Immunol Immunopathol. 2005; 106:259–267.

11. Moeini H, Omar AR, Rahim RA, Yusoff K. Development of a DNA vaccine against chicken anemia virus by using a bicistronic vector expressing VP1 and VP2 proteins of CAV. Comp Immunol Microbiol Infect Dis. 2011; 34:227–236.

12. Moeini H, Omar AR, Rahim RA, Yusoff K. Improving the potency of DNA vaccine against chicken anemia virus (CAV) by fusing VP1 protein of CAV to Marek's disease virus (MDV) type-1 VP22 protein. Virol J. 2011; 8:119.

13. Morgan RW, Gelb J Jr, Schreurs CS, Lütticken D, Rosenberger JK, Sondermeijer PJ. Protection of chickens from Newcastle and Marek's diseases with a recombinant herpesvirus of turkeys vaccine expressing the Newcastle disease virus fusion protein. Avian Dis. 1992; 36:858–870.

14. Nagai Y, Klenk HD, Rott R. Proteolytic cleavage of the viral glycoproteins and its significance for the virulence of Newcastle disease virus. Virology. 1976; 72:494–508.

15. Nakaya T, Cros J, Park MS, Nakaya Y, Zheng H, Sagrera A, Villar E, García-Sastre A, Palese P. Recombinant Newcastle disease virus as a vaccine vector. J Virol. 2001; 75:11868–11873.

16. Pedersen N, Hansen S, Heydenreich AV, Kristensen HG, Poulsen HS. Solid lipid nanoparticles can effectively bind DNA, streptavidin and biotinylated ligands. Eur J Pharm Biopharm. 2006; 62:155–162.

17. Rajawat YS, Sundaresan NR, Ravindra PV, Kantaraja C, Ratta B, Sudhagar M, Rai A, Saxena VK, Palia SK, Tiwari AK. Immune responses induced by DNA vaccines encoding Newcastle virus haemagglutinin and/or fusion proteins in maternal antibody-positive commercial broiler chicken. Br Poult Sci. 2008; 49:111–117.

18. Reddy GS, Srinivasan VA. Use of BHK cell culture-adapted Newcastle disease virus for immunization of chicks. Vaccine. 1992; 10:164–166.

19. Rees S, Coote J, Stables J, Goodson S, Harris S, Lee MG. Bicistronic vector for the creation of stable mammalian cell lines that predisposes all antibiotic-resistant cells to express recombinant protein. Biotechniques. 1996; 20:102–104. 106108110

20. Reynolds DL, Maraqa AD. Protective immunity against Newcastle disease: the role of antibodies specific to Newcastle disease virus polypeptides. Avian Dis. 2000; 44:138–144.

21. Reynolds DL, Maraqa AD. Protective immunity against Newcastle disease: the role of cell-mediated immunity. Avian Dis. 2000; 44:145–154.

22. Richter E, Wick G. Fluoro-immuno-cytoadherence (FICA): a new method for the identification and enumeration of antigen-binding cells. Z Immunitatsforsch Immunobiol. 1977; 152:351–362.

23. Sakaguchi M, Nakamura H, Sonoda K, Hamada F, Hirai K. Protection of chickens from Newcastle disease by vaccination with a linear plasmid DNA expressing the F protein of Newcastle disease virus. Vaccine. 1996; 14:747–752.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download