Introduction
Japanese encephalitis (JE) is a mosquito-transmitted viral disease caused by JE virus (JEV) belonging to genus Flavivirus of the Flaviviridae family. JE is principally a disease occurring in rural agricultural areas where vector mosquitoes proliferate in close association with pigs, wading birds, and ducks. Horses and humans are the dead-end hosts, in which JEV causes acute encephalitis. JE occurs only sporadically in horses with low morbidity (0.045~0.3%) and fatality rates ranging between 5 and 30% [19,41]. The virus may infect a number of other domestic animals, including cattle, sheep, goats, dogs, and cats, in which these infections are typically asymptomatic.
JE is prevalent in Southeast Asia and has also been reported in Indonesia, northern Australia, Papua New Guinea, and Pakistan [11]. Approximately, 3 billion people live in JEV-endemic areas where 50,000 cases and 15,000 human deaths due to JE are reported annually [11]. Seasonal outbreaks of JE also occur regularly in human population in several parts of India [8,10,27]. In Haryana (India), JE epidemics among humans were first reported in 1990 [34]. Subsequently, human cases have been reported regularly in this state [17,18,31].
The JEV genome is encoded by a plus-sense, single-stranded RNA of approximately 11 kb [1]. It has a single open reading frame that codes for a polyprotein of 3,432 amino acids, which is subsequently cleaved into three structural [capsid (C), premembrane (prM), and envelope (E)] and seven non-structural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) proteins [1]. The E protein is the most important structural protein and induces the production of virus-neutralizing (VN) antibodies which impart protective immunity against JEV [1]. Based on E gene phylogenetic analysis, JEV strains have been classified into five genotypes (GI-V) [25,32,35,37,40]. Genotype I (GI) includes strains isolated in Southeast Asia, Australia, northern Thailand, Cambodia, Korea, Japan, and India. Genotype II (GII) includes strains isolated in southern Thailand, Malaysia, Indonesia, and Australia. Genotype III (GIII) includes strains isolated in Southeast Asia, Japan, China, Korea, Taiwan, and the Central Asian sub-continent including India [27,32,37]. GIV includes strains isolated in Indonesia alone and GV includes a single strain isolated in Singapore [10,14,25,37]. GIII is widely distributed throughout Asian countries and most of the JEV strains isolated in India belong to GIII [27,37]. JEV GI has recently been reported in human patients from Gorakhpur (Uttar Pradesh, India) [10].
Sporadic clinical cases of JE in horses have been reported in various countries including Japan [19,41], Hong Kong [20], Taiwan [21], and India [31]. In addition, JE seropositivity among equines has been reported in Nepal, Korea, Indonesia, and India [2,9,13,23,26,28,31,33,39]. However, there is no report of JEV isolation from equines in India. Resurgence of JE cases in human populations in India during the last decade [8,27] highlights the need for JEV surveillance in horses. In this paper, we report evidence of JEV infection in horses in Haryana (India) and the first isolation of JEV from a horse.
Materials and Methods
Collection of samples
Serum samples from apparently health horses in 11 districts (Ambala, Fatehabad, Gurgagon, Hisar, Jhjjar, Jind, Karnal, Panchkula, Rohtak, Sirsa and Yamunanagar) of Haryana state (India) were collected between 2006 and 2010 (Table 1). None of the horses had been vaccinated against JEV. Blood samples from jugular vein were collected and transported to the laboratory at 4℃. Serum was separated by centrifugation at 1,000 × g for 5 min and stored at -30℃ until further use.
Blood (with EDTA) and serum samples from two horses in a farm exhibiting neurological signs (circling, pedaling followed by paralysis and recumbency) were collected aseptically for JEV isolation and RT-PCR.
Brain tissues and cerebrospinal fluid from two dead horses were collected at post-mortem and transported to the laboratory on ice. Paired serum samples were also collected from other 37 apparently healthy horses from the same equine farm.
Resting mosquitoes found around the equine farm were collected using mosquito cages measuring approximately 30 × 30 × 30 cm as previously described [7]. Briefly, mosquitoes were collected by a series of quick forward, backward, up, and down sweeping movements of the cage through vegetation and bushes for 2 min to disturb the resting mosquitoes. This was done at least 10 times for 1 h after dusk. The mosquitoes were stored at 4℃ until engorged females of Culex spp. were identified microscopically and isolated. Female mosquitoes were pooled into six groups, each containing 100~300 mosquitoes. Homogenate (10% w/v) of each pool of mosquito was made in Eagle's minimal essential medium (EMEM; Sigma-Aldrich, USA) containing 2% fetal bovine serum (FBS; Sigma-Aldrich, USA) using pestle and mortar. The homogenate was centrifuged at 12,000 × g for 5 min and the supernatant was stored at -30℃.
Viruses and cells
A JEV strain (P20778; JEV GIII strain isolated from a human patient at Vellore, India) procured from the National Institute of Virology, Pune (India) was used to prepare antigen for a hemagglutination inhibition (HI) test. The virus was propagated in a-day old Swiss Albino suckling mice by intra-cerebral inoculation (20 µL of JEV inoculum per mouse) as described [6]. For the virus neutralization test (VNT), the virus was cultured in porcine stable kidney (PS) cells obtained from the National Centre for Cell Sciences, Pune (India). The cells were grown at 37℃ in EMEM supplemented with 10% FBS, 100 IU/mL penicillin, 100 µg/mL streptomycin and 0.25 µg/mL amphotericin-B (Sigma-Aldrich, USA).
Hemagglutination inhibition test
The HI test was carried out in duplicate in 96-well microplate (Tarson, India) with using 8 HA units/25 µL of antigen prepared from the brains of suckling mice infected with JEV P20778 strain following sucrose-acetone extraction method [6]. Serum samples were treated with cold acetone to remove non-specific inhibitors and were adsorbed with goose erythrocytes as described [6]. Hyper-immune serum against JEV (raised previously in rabbits by immunization with inactivated JEV vaccine, procured from Central Research Instititute Kasauli, India) was used as a positive control. HI titers of 1 : 20 and above were considered positive.
Virus neutralization test
Serial two-fold dilutions of JEV strain (P20778) (four wells per dilution per virus) in 100 µL volume of EMEM supplemented with 2% FBS were made in 96-well culture plates (Costar, USA). To each well, 100 µL of PS cells (2×105 cells/mL in EMEM supplemented with 10% FBS) were added and incubated for 5 days at 37℃ in 5% CO2. The wells showing cytopathic effects (CPE) were recorded to calculate 50% tissue culture infectivity dose (TCID50) of the virus. At passage 5, the virus titer was 107 TCID50/mL. VNT was performed in 96-well tissue culture plates using PS cells. Serum samples were heat inactivated at 56℃ for 30 min and serial two-fold dilutions of the serum in a 50 µL volume was mixed with an equal volume of 300 TCID50 of the JEV or West Nile virus (WNV). After incubating the virus-serum mixture at 37℃ for 1 h, 100 µL of a PS cell suspension (2 × 105 cells/mL) was added to each well. The plates were incubated at 37℃ in 5% CO2 for 5 days and CPE in each well was recorded. Neutralization titers were expressed as the reciprocal of the highest serum dilution that inhibited CPE in 50% of the cultures.
Monoclonal antibody-based capture ELISA
The ELISA was standardized following the method of Johnson et al. [16] with minor modifications. Briefly, ELISA modules (Greiner, USA) were coated overnight at 4℃ with EJC7, a JEV-specific murine monoclonal antibody raised in our laboratory, at a dilution of 1 : 8,000 in carbonate buffer (pH 9.6). The plates were blocked with 3% bovine serum albumin fraction V (BSA; Sigma Aldrich, USA) for 1 h at 37℃. After washing five times with phosphate buffer saline (PBS) containing 0.05% Tween 20 (PBS-T), 50 µL of formalin-inactivated JEV strain (P20778) grown in PS cells were added and incubated at 37℃ for 1 h. After five washes with PBS-T, test horse serum (50 µL) diluted 1 : 50 in 3% BSA was added to duplicate wells. Hyper-immune serum against JEV raised previously in rabbits was used as positive control and normal rabbit serum as negative control. After incubating for 1 h at 37℃, the plates were washed five times with PBS-T and 50 µL of goat anti-horse HRP (Sigma-Aldrich, USA) was added at a dilution of 1 : 2,000. The plates were incubated for 1 h at 37℃ and then washed five times with PBS-T. Fifty microliters of tetramethylbenzidine substrate (Sigma-Aldrich, USA) was added. The plates were incubated for 10 min and the reactions were stopped by adding 50 µL of 2.5 M sulfuric acid to each well. The absorbance was measured at 450 nm in an ELISA reader (Biotek, USA). The cut-off value of 0.15 was determined by the mean absorbance of the known negative serum samples plus three standard deviations.
Virus isolation and identification
Leukocytes were separated from 0.5 mL blood (with EDTA) using histopaque-1077 (Sigma-Aldrich, USA) and lysed in EMEM containing 2% FBS by two cycles of freezing and thawing. Samples of the leukocyte lysates and serum were filtered through a 0.2 µm membrane (Millipore, USA) and 20 µL aliquots of each sample were used to intra-cerebrally inoculate a litter of at least six 1-day old Swiss albino suckling mice, procured from Disease-free Small Animal Facility, Haryana Agricultural University, Hisar (India). Two mice from the same litter were inoculated with EMEM containing 2% FBS as a negative control. The mice were observed for 14 days to monitor the development of JE-specific clinical signs.
The samples of the leukocyte lysates and serum were also inoculated in duplicate on PS cell monolayers grown in 25 cm2 tissue culture flasks (0.5 mL volume per flask). After adsorption for 1 h at 37℃, the cell monolayers were washed once with PBS before adding EMEM supplemented with 2% FBS. The cells were incubated at 37℃ in CO2 incubator for 5 days or until CPE was observed. If no CPE was observed, the cells were frozen and thawed three times and the cell culture lysate passaged in fresh PS cells at least 5 times, before determining that the sample was negative. The identity of the isolated virus was confirmed by RT-PCR and a VNT using varying two-fold dilutions of the virus with a known concentration of hyper-immune serum (HIS) raised against JEV in rabbits.
RT-PCR
Viral RNA from 100 mg of brain tissue or 200 µL of cell culture lysate or serum samples was extracted using TRI reagent (Sigma-Aldrich, USA) according to the manufacturer's instructions. Viral RNA was reverse-transcribed using Stratascript reverse transcriptase (Stratagene, USA) and random primers at 42℃ for 1 h in a final volume of 25 µL. PCR-amplificatin was done using primers designed to amplify a 146 bp region of the 3'NTR region of the JEV genome spanning nucleotide 10,726 to 10,871. PCR amplification was done with 3 µL of cDNA and 22 µL of a PCR mixture containing 1 × Taq buffer, 500 nM of each primer, 200 µM of a dNTP mix, 2.0 mM MgCl2, and one unit of Taq DNA polymerase (Sigma-Aldrich, USA). The thermal cycling conditions consisted of an initial denaturation at 95℃ for 5 min followed by 30 cycles each of 95℃ for 1 min, 51℃ for 30 sec, 72℃ for 30 sec, and a final elongation step at 72℃ for 5 min. PCR amplification was also done using E gene primers as described [5]. RT-PCR products (5 µL) were separated by electrophoresis in a 2% agarose gel at 90 V for 1 h. PCR amplicons of the 3'NTR (expected size, 146 bp) and E gene (expected size, 291 bp) were visualized under gel documentation system (Syngene, UK).
Cloning and nucleotide sequencing
The synthetic oligonucleotides used for sequencing of 2,500 nuceleotides of JEV genome were: forward primer ATCAATATGCTGAAACGCGGTC and reverse primer TTCCTTGTGCGCTTTGTGGACGA. Viral RNA from JEV-infected PS cells was reverse-transcribed using reverse primer and Stratascript reverse transcriptase. cDNA copies were generated by PCR using Dream Taq PCR mix (Fermentas, Germany). The PCR products were ligated into a pGEMT-Easy vector (Promega, USA) and transformed into E. coli JM109 cells (Promega, USA) using Transform-aid bacterial transformation kit (Fermentas, Germany) as per manufacturer's protocol. Positive transformants were selected by colony PCR with the above mentioned primers and restriction enzyme digestion of recombinant plasmid with EcoRI. Double-stranded DNA sequencing of the recombinant clone was performed by primer-walking at the National Facility for DNA Sequencing, University of Delhi South Campus, India and Sequence Analyzer 3730 (Applied Biosystems, USA).
Nucleotide sequence of the equine JEV isolate JEV/eq/India/H225/2009 (accession No. HQ018880) was analyzed using the BLAST database (NCBI, USA). The nucleotide sequences were aligned using ClustalW and a phylogenetic tree generated by the maximum parsimony method using MEGA 4.0 software [36]. Statistical analysis of the tree was performed by bootstrap re-sampling (1,000 data sets) of the multiple alignments.
Results
JEV seroprevalence
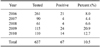
Horse serum samples were analyzed by HI and VNT to determine JEV seroprevalence. Out of 637 equines in the state of Haryana tested during 2006 and 2010, 67 (10.5%) were positive for JEV antibodies according to both HI and VNT (Table 1). Chi-square-analysis indicated that there was no significant difference in year-to-year (p > 0.05) or district-to-district (p > 0.05) prevalence of JEV among the horses.
Clinical evidence of JEV in equines
Two male Marwari foals less than 1 year old kept at an equine farm in Hisar (Haryana) developed ataxia with extended hind legs, anorexia, and became recumbent. One foal (No. H225) became ill during the first week of March 2009, remained sick for 2 weeks, and then died. The other foal (No. H238) exhibited similar clinical signs in August 2009 and died within 2 days. Post-mortem examination of the first foal (No. H225) revealed widespread congestion of meninges, cerebral hemispheres, and cerebellum. Histopathological findings included widespread congestion of blood vessels along with swelling of endothelial cells in sections of the cerebral hemispheres. Multi-focal areas of neuronal swelling, degeneration, and necrosis could be seen along with glial cell proliferation. Similar post-mortem lesions and histopathological changes, but to a lesser extent, were observed in the brain of second foal (No. H238). This foal also had a concurrent Trypanosoma evansi infection as revealed by microscopic examination of a blood smear.
Virus isolation and identification
Blood and serum samples were collected on the first day of illness and daily afterwards until the foals died. Serum samples of the foal (No. H225) collected on day 2 and 3 of illness and that of foal H238 collected on the second day of illness were positive for Flavivirus RNA as revealed by amplification of a 146 bp fragment of 3'NTR by RT-PCR (Fig. 1A). Furthermore, the presence of JEV was confirmed by RT-PCR through amplification of a 291 bp E gene fragment (Fig. 1B). Serum and blood samples collected from foal H225 during day 3~14 of illness were negative for Flavivirus and JEV RNA.
Samples from both the foals were used to intracerebrally inoculate suckling mice and PS cells for virus isolation. Serum sample from foal H225 collected on the second day of illness produced typical nervous symptoms in mice including hyper-excitement followed by ataxia, walking with a wide stance, recumbency, leg pedaling, paralysis, and death 10 days post-inoculation (dpi). After the second passage in mice, nervous symptoms developed within 4~5 days. The serum as well as leukocyte lysate from foal H225 produced characteristic CPE in PS cells during the second passage on 4 dpi.
Virus isolation was confirmed by VNT using HIS against JEV raised in rabbits. The virus passaged in PS cells was neutralized by rabbit serum at a dilution of 1 : 1,600 at passage 2. Virus identity was also confirmed by RT-PCR amplification of a partial 3'NTR gene (146 bp, Fig. 1A) and E gene (291 bp, Fig. 1B). The isolated strain was named JEV/eq/India/H225/2009. JEV was not isolated from the second foal (No. H238) even after five passages in PS cells.
Serum samples collected from these two foals were tested for JEV antibodies. HI and VNT failed to detect antibodies against JEV in the serum samples from foal H225 until day 14 of illness. However, monoclonal antibody-based capture ELISA demonstrated the development of JEV antibodies in the serum of foal H225 from day 10~14 of illness. The other foal (No. H238) that died within 2 days of illness was negative for JEV antibodies according to all three assays. Paired serum samples from all the equines (n = 37) on the farm were tested by HI and were negative for JEV antibodies in August 2009. On subsequent testing in May 2010, 10 out of 37 were positive for JEV antibodies (HI titers between 40 and 80) and later became seronegative by August 2010.
Mosquito screening
Homogenates from six pools of female Culex spp. mosquitoes collected from vegetation around the equine farm during August 2009 were analyzed by RT-PCR. One pool of mosquitoes was positive for JEV RNA that yielded a 291 bp product from E gene-based PCR (data not shown). However, attempts to isolate virus from this mosquito pool in suckling mice or PS cells were not successful.
JEV sequence analysis
A partial genome (2,500 nucleotides) including the structural gene region of JEV isolate (JEV/eq/India/H225/2009) was sequenced in the present study (accession No. HQ018880). On BLAST, the JEV/eq/India/H225/2009 genome resembled those of other known JEV strains showing a 99% identity with nucleotides 134~2,633 of the Vellore strain (accession No. AF080251) isolated from India in 1956. Additionally, the strain isolated in the present study had more than 97% nucleotide identity with different JEVs isolated in India, and a 97% identity with the Nakayama strain (accession No. EF571853).
Phylogenetic analysis of 1,500 nucleotides from the E gene sequence (accession No. GQ387646) of the JEV/eq/India/H225/2009 isolate was performed with gene sequences of 22 known JEV strains from different geographical locations (Table 2). The horse isolate (JEV/eq/India/H225/2009) grouped with GIII and showed a nucleotide sequence identity of more than 98% with JEV isolates from southern India (Fig. 2A) such as the Vellore (G8924 and P20778) or Chennai isolate (782219). Phylogenetic analysis of JEV/eq/India/H225/2009 based on a 240-bp nucleotide sequence of the C/prM region was also done for 20 JEV strains, including isolates from India (Fig. 2B). The prM gene phylogenetic tree also revealed that the JEV/eq/India/H225/2009 isolate was grouped with GIII.
Discussion
JE remains endemo-epidemic in several Asian countries. Several outbreaks are reported every year from different regions in humans as well as domestic animals. In northern India, large JE epidemics occur in humans during the summers when vector activity is abundant whereas JE tends to be endemic throughout the year in the southern region of the country [24]. Human JE cases have also been reported sporadically in the northwestern state of Haryana [17,18,29,30,34], indicating that JEV is circulating in this region.
JEV seroprevalence of 10.8% as reported by this paper in horses from 11 out of 19 districts of Haryana between 2006 and 2010 also indicates that JE is enzootic among equines in Haryana. Sporadic cases of JE in horses have been observed in Japan, Hong Kong, and Taiwan despite the use of JEV vaccinations in these countries [15,20,21,41]. In India, JEV vaccinations are not administered to horses and a focal outbreak occurred in four horses during 1999 in the western state of Maharashtra [31]. Serological evidences of JE infection in domestic animals from other states in India have been previously reported [2,9,13,23,28,33].
Two foals showing neurological signs were detected positive for Flavivirus infection by RT-PCR based on 3'NTR primers, which amplify a 146 bp fragment from both JEV and WNV. Further testing by E gene-based RT-PCR confirmed that two foals and one pool of mosquito from the same equine farm were specifically positive for JEV.
A definite diagnosis of JEV by virus isolation is indisputable but rare [41]. Attempts to isolate JEV from horses have not always been successful [20,21,31]. However, we isolated JEV from the serum of a foal collected on day 2 of illness. Virus could not be isolated from samples collected on other days other than day 2 of illness. This was probably due to the fact that viremia titers in equines are very low and transient, detected only between the first and fourth day of infection [12]. JEV was not isolated from the other foal (No. H238), which died on day 2 of illness. It is worth mentioning that this foal had a concomitant acute Trypanosoma evansi infection.
Antibodies against JEV in equines appears after 1~2 weeks of infection and can be detected by various serological assays [6,9,15,19,21,28]. In the present study, HI and VNT failed to detect antibodies in JEV-infected foals until day 14 of illness. However, capture ELISA, which appears to be more sensitive than HI and VNT, detected antibodies in the serum samples collected from day 10~14 of illness.
E gene phylogenetic analysis indicated that JEV/eq/India/H225/2009 was GIII and clustered more closely with the Vellore group of isolates. The JEV/eq/India/H225/2009 had more than 99% nucleotide identity with JEV G8924, a Vellore mosquito isolate recovered in 1956. It has been reported that genetic diversity exists among GIII JEV strains from India [37]. Based on E gene nucleotide divergence (cut-off value of 3.4%), Indian GIII strains may be grouped into four different clusters [27,37]: the Vellore group (represented by P20778 and G8924 strains), Gorakhpur-2005 group (GP-82-like strains from the 2005 Gorakhpur JEV outbreak), Bankura group (represented by GP78 and 733913 strains), and Nepal group (represented by 7812474 from Assam and B2524 strain from Nepal).
Phylogenetic analysis of the C/prM region places the horse JEV isolate into GIII [3,4,22,38]. However, unlike in the E gene-based tree, no significant clustering pattern was observed within the GIII Indian JEV strains in this study. This indicates that the C/prM region can be used only for genotyping and is not an ideal region for determining phylogenetic relationships among closely related GIII strains [37].
There are only a few reports of isolation and genotyping JEV isolates from horses [20,21,32,41]. Based on E gene sequence phylogeny, a virus strain (Tottori) isolated from the cerebrum of a half-bred horse kept in the Tottori Prefecture, Japan was classified as GI [41]. Additionally, a JEV isolate (V304) from a thoroughbred horse in Hong Kong was classified as GII [20] and an equine JEV isolate of 1947 from Japan as GIII [32]. Based on C/prM gene sequences, equine isolates from two horses in Taiwan (H1 and H2) were typed as GIII [21].
In conclusion, this is the first report of JEV isolation from a horse in India. Our results from E and C/prM gene sequence analysis clearly showed that the first equine JEV isolate from India belongs to the GIII genotype. These findings indicated that JEV GIII belonging to the Vellore group lineage is circulating in this region. JEV RNA was also detected in Culex spp. mosquitoes collected from the same farm housing a symptomatic horse. It is worth mentioning that Culex tritaeniorhynchus is the most important vector for JEV transmission in India [7]. Evidence of JEV in this region (in equines and mosquitoes) may have potential implications for future spread of JEV in humans and other animal populations. Additional surveys in animals, humans, and vector populations in Haryana (India) are necessary to better understand JEV epidemiology.




 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download