Abstract
The purpose of this study was to determine whether manually plucked hairs might serve as an alternative sample for a quantitative real time polymerase chain reaction (qRT-PCR) testing. Twenty three, 1~3 week old, non-bovine viral diarrhea virus (BVDV) vaccinated calves, found to be positive for BVDV by immunohistochemical staining, were selected and hairs were manually plucked from the ear. qRT-PCR was performed on samples consisting of more than 30 hairs (30~100) and whole blood. All 23 animals were positive for the virus by qRT-PCR performed on the whole blood and when samples of more than 30 hairs were assayed. Additionally, qRT-PCR was performed on groups of 10 and 20 hairs harvested from 7 out of 23 immunohistochemical staining-positive calves. When groups of 20 and 10 hairs were tested, 6 and 4 animals, respectively, were positive for the virus.
Bovine viral diarrhea virus (BVDV) infection is an important disease of cattle, leading to reproductive failure, decreased production, morbidity and mortality in cattle and other artiodactylids, including sheep, goats, bison; it also affects a variety of cervids, including elk, eland, mouse deer, as well as white tailed, mule, red, roe, and fallow deer [5]. A test-and-cull approach, combined with farm biosecurity measures, resulted in the successful eradication of BVDV infection in Scandinavia [4]. The efficacy of the test-and-cull approach relies heavily on the sensitivity and specificity of the available assays, as well as on compliance, which is influenced by the cost and ease of sampling and shipping. However, some form of limitation exists with these currently available tests [2,3]. We investigated the value of plucked hairs as an appropriate sample for BVDV screening, based on the epithelial distribution of this virus, as visualized by immunohistochemistry (IHC). Typically, large numbers of positive staining keratinocytes are present within the skin of persistently infected cattle (Fig. 1). Most of the epithelial portion of the hair follicle (bulb) remains with the hair after plucking suggesting that plucked hairs may represent a feasible and convenient sample for BVDV detection.
Samples from 29 calves were utilized in this study. Twenty three calves were BVDV positive and 6 calves were BVDV negative. A full thickness section of the mid to distal ear pinna, measuring 5~10 mm in diameter, was collected from these 29 calves, placed in 10% neutral buffered formalin (ear notch) and submitted to the laboratory for routine screening for BVDV. In addition, approximately 3~4 mL blood were also collected in tubes containing EDTA and hair samples with hair follicles (bulb). Ear notches were processed for routine histopathology and then subjected to IHC. Briefly, 3~4 µm sections were mounted on positively charged slides and dried at 55~72℃. Tissue sections were then deparaffinized in xylene and rehydrated through decreasing concentrations of ethanol and finally water. Endogenous peroxidase activity was quenched by a solution of 3% hydrogen peroxide in methanol for 15 min, after which the sections were washed, digested in 0.1% protease for 20 min at 37℃, and washed again. The primary antibody was a monoclonal antibody to BVDV (15.c.5; Cornell University, USA). This antibody was diluted 1 : 50 in a commercial diluent (BioGenex, USA) and incubated with the tissue sections for 30 min at room temperature, using an automated immunostainer (i6000; BioGenex, USA), followed by incubation with an amplifier (Super Enhancer; BioGenex, USA) and casein-based blocking solution (Super Block; BioGenex, USA) at room temperature for 20 min, after which they were washed. The sections were incubated for 30 min at room temperature with a secondary antibody conjugated to a horseradish peroxidase polymer (Polymer HRP reagent; BioGenex, USA), washed, then incubated with NovaRED (Vector, USA) for 10 min. Finally, the tissue sections were washed, counterstained with Mayer's hematoxylin for 1 min, and then rinsed, dehydrated on an automatic stainer (Tissue-Tek DRS; Sakura Finetek, USA), cleared, and mounted on an automatic coverslipper. With each sample, a positive control tissue (i.e. an ear notch sample previously determined to be positive by immunohistochemical analysis) was included. Slides with test and positive control tissue were prepared in duplicate, with one treated as described above and the other treated as above, but without the addition of primary antibody.
Twenty three, 1~3 week old, non-BVDV vaccinated, Holstein Friesian and mixed breed calves that tested positive twice by immunohistochemical staining were selected for further evaluation. Hair shafts with roots were manually plucked from the ear of each of these calves and placed in 1 mL of RNA stabilization solution (RNAlater; Ambion, USA). Approximately 3~4 mL of blood was also collected into EDTA for white blood cell (WBC) purification. Both samples were transported to the laboratory at ambient temperature for BVDV 1 and 2 qRT-PCR.
Hair sample groups were prepared for qRT-PCR by manually grinding the samples in 200 µL of RNA stabilization solution supplemented with buffer (RLT buffer, RNeasy Mini Kit; Qiagen, USA) containing 2β-mercaptoethanol, using a Kontes pellet pestle. Total RNA purification was performed using an RNA isolation kit per manufacturer's protocol, with a final elution volume of 30 µL. The WBC samples were processed for total RNA extraction by using a kit designed for extraction of RNA from fresh whole blood (WBC QIAamp RNA Blood Mini Kit; Qiagen, USA). According to a previously published protocol [1], BVDV qRT-PCR was performed using primers and probes for BVDV type 1 and type 2 discrimination, specifically, a commercial master mix (RT-PCR Master Mix; Eurogentec, USA) was used with the addition of 10 µM primers and 1 µM probes for a total reaction volume of 25 µL. Reaction parameters were carried out in a thermocycler (SmartCycler II; Cepheid, USA), as follows: 1 cycle each of 48℃ for 30 min then 95℃ for 10 min and subsequently 40 cycles of 95℃ for 15 sec with 58℃ for 60 sec. Positive controls consisted of RNA purified as above from either cell culture propagated BVDV type 1 (Singer strain) and type 2 (strain 125) for hair samples, or BVDV (type 1 and 2) infected WBC obtained from persistently infected calves. Negative preparation controls were extractions made with only kit reagents and produced at the time of sample extractions. In particular, negative PCR controls consisted of the master mix without template. Samples with Ct values ≤ 35 were considered positive for the presence of the BVDV genome, Ct values 35.1~40 were considered suspect and Ct values > 40 were considered not to contain detectable amounts of viral RNA.
All 23 animals that tested positive for BVDV by IHC were also positive for either BVDV-1 or BVDV-2 by qRT-PCR, when performed on at least 30 (30~100) plucked hairs and on WBC preparation. Thus, when qRT-PCR was performed on hairs, twenty two of these animals tested positive for BVDV-1 and one tested positive for BVDV-2. Blood was not available for processing from this calf (Table 1). Twenty two BVDV-1-infected animals were also positive when qRT-PCR was performed using their respective WBC sample.
In order to surmise how many hairs per sample may be needed to obtain an adequate sensitivity, qRT-PCR was performed on 3 different quantities of hairs from seven of the 23 animals. Hair samples were therefore separated into groups consisting of 10, 20 and more than 30 (30~100) individual hairs. The 7 animals were selected as a convenience sample number that was manageable within the budget of the study. Because pooling is a helpful way to reduce the expenditure required to obtain results of screening tests, smaller numbers of hairs were tested in seven of the animals.When groups consisting of 20 and 10 hairs were assayed, only six and four of the seven samples were positive, respectively (Table 2).
The results from this study indicate that using plucked hairs for qRT-PCR testing is a viable alternative to WBC samples for this assay or to using ear notches for analysis by IHC. Complete agreement between the results of the IHC performed on the ear notches and qRT-PCR performed on samples consisting of at least 30 plucked hairs suggests a strong association between the two tests, even with the relatively small number of animals tested in our study. However, this test also has few limitations, because its sensitivity and specificity remains unknown and it is expansive. Therefore, in order to establish the sensitivity and specificity of our method, a study consisting of a larger number of animals is underway.
qRT-PCR performed using plucked hairs offers many advantages. Hair samples are easy and inexpensive to collect, do not utilize specialized sampling equipment and have simple storage and shipping requirements. Further, this test is rapid compared to other available tests, including IHC and ELISA. The use of formalin is also eliminated, reducing hazards associated with personal exposures, as well as the need for hazardous waste disposal. These advantages, together with the work described here, indicate that the use of hair samples may be a viable alternative to the use of either ear notches or blood samples.
Figures and Tables
Fig. 1
Multiple hair follicular cells were positive for bovine viral diarrhea virus (BVDV) antigen using immunohistochemistry. Note the positive staining in epidermal cells (E), hair (H), follicular cells (F), and sebaceous glands (S).
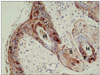
Table 1
Comparison of results of immunohistochemical staining on ear notch samples, qRT-PCR for bovine viral diarrhea (BVD)-1 and BVD-2 on buffy coat samples, and on samples of at least 30 plucked hairs

All calves except one (number 22) were positive for BVD-1. Blood was not available from this calf. The values in parenthesis are Ct values of samples. Ct values ≤ 35 were considered positive for the presence of the BVDV genome, Ct values 35.1~40 were considered suspect and Ct values > 40 were considered not to contain detectable amounts of viral RNA. NA: Blood was not available.
References
1. Bhudevi B, Weinstock D. Fluorogenic RT-PCR assay (TaqMan) for detection and classification of bovine viral diarrhea virus. Vet Microbiol. 2001. 83:1–10.

2. Cornish TE, van Olphen AL, Cavender JL, Edwards JM, Jaeger PT, Vieyra LL, Woodard LF, Miller DR, O'Toole D. Comparison of ear notch immunohistochemistry, ear notch antigen-capture ELISA, and buffy coat virus isolation for detection of calves persistently infected with bovine viral diarrhea virus. J Vet Diagn Invest. 2005. 17:110–117.

3. Driskell EA, Ridpath JF. A survey of bovine viral diarrhea virus testing in diagnostic laboratories in the United States from 2004 to 2005. J Vet Diagn Invest. 2006. 18:600–605.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download