Abstract
Background
Cordyceps militaris has been used in traditional medicine to treat numerous diseases and has been reported to possess both antitumor and immunomodulatory activities in vitro and in vivo. However, the pharmacological and biochemical mechanisms of Cordyceps militaris extract (CME) on macrophages have not been clearly elucidated. In the present study, we examined how CME induces the production of proinflammatory cytokines, transcription factor, and the expression of co-stimulatory molecules.
Methods
We confirmed the mRNA and protein levels of proinflammatory cytokines through RT-PCR and western blot analysis, followed by a FACS analysis for surface molecules.
Results
CME dose dependently increased the production of NO and proinflammatory cytokines such as IL-1β, IL-6, TNF-α, and PGE2, and it induced the protein levels of iNOS, COX-2, and proinflammatory cytokines in a concentration-dependent manner, as determined by western blot and RT-PCR analysis, respectively. The expression of co-stimulatory molecules such as ICAM-1, B7-1, and B7-2 was also enhanced by CME. Furthermore, the activation of the nuclear transcription factor, NF-κB in macrophages was stimulated by CME.
Cordyceps militaris is also known as the rare Chinese caterpillar fungus (1) and has benefits for the human body including effects on the circulatory, immune, respiratory, and glandular systems. It is commonly used in the orient to replenish the kidneys and soothe the lung as well as in the treatment of hyperglycemia, hyperlipidemia (2,3), renal dysfunction, and liver disease (4); it is also thought to have anti-mutagenic (5) and anti-angiogenesis (6) capabilities. Recently, several studies demonstrated that extracts of Cordyceps militaris had multiple pharmacological actions which included anti-inflammatory activity (6), improvement of insulin resistance and insulin secretion (2), and tumor suppression activity. Addition, it possesses antioxidant activity to a greater extent than Cordyceps sinensis or Cordyceps kyushuensis (7). Because natural Cordyceps militaris is rare and expensive, many scientists have examined its life cycle with the aim of developing techniques for the isolation of fermentable strains. Even though the immunomodulatory role of CME in immune response is being explored and further elucidated, the regulatory effects of CME on specific functions of macrophages has yet to be fully explained.
Macrophages, a type of differentiated tissue cell which originate as blood monocytes, play an important role in immune and allergic reactions as well as in inflammation (8). These cells have several functions including the killing and removal of pathogenic microbes and the processing and presentation of antigens which have been ingested by lymphocytes (9). Macrophages are essential cellular components of the body's host defense system, playing critical roles in both innate and adaptive immunity. Also, these cells collaborate extensively with T cells through cell-cell and cytokine-mediated interactions to coordinate the evolution of inflammatory responses. In response to microbes and their products, such as lipopolysaccharide (LPS), macrophages secrete various inflammatory cytokines including interleukin (IL)-1β, IL-6, IL-12, and tumor necrosis factor (TNF)-α through the activation of nuclear factor (NF)-κB; they also express NF-κB-dependent inducible nitric oxide (NO) synthase (iNOS) (10,11) and cyclooxygenase-2 (COX-2) (12). Many diseases, such as rheumatoid arthritis (RA), arteriosclerosis, chronic hepatitis, and pulmonary fibrosis (13-16) involve the overproduction of inflammatory mediators. Although large amounts of macrophage-derived proinflammatory mediators can cause several immune diseases, proper regulation of macrophage function by immunomodulating agents could help protect the host from various pathogenic and cancerous attacks.
Nuclear factor-κB, a nuclear transcription factor, regulates the expression of various genes, including IL-1β, TNF-α, iNOS, and COX-2, that play critical roles in apoptosis, tumorigenesis, various autoimmune diseases, and inflammation (17,18). Because of its ubiquitous role in the pathogenesis of inflammatory gene expression, NF-κB is currently a target for treating various diseases (19). Nuclear factor-κB is activated in response to various inflammatory stimuli, including bacterial LPS, cytokines, and viral proteins (20). Under normal conditions, NF-κB is present in the cytoplasm as an inactive hetero-trimer consisting of p50, p65, and IκBα subunits. Upon activation, degradation of IκBα exposes a localization signal on the p50/p65 hetero-dimer leading to nuclear translocation and to its binding to a specific sequence in several promoters which, in turn results in the transcription of proinflammatory genes.
Given that these effects have been suggested to play an important role in disease therapy, we investigated whether CME could increase the production of proinflammatory mediators in macrophages.
Lipopolysaccharide (LPS) was purchased from Sigma (St. Louis, USA). Cell culture media DMEM, antibiotic-penicillin/streptomycin solution, and fetal bovine serum (Hyclone, Logan, USA) were used for the cell culture.
Cultures of fruiting bodies of Cordyceps militaris were supplied by CM Biotec, Kangnung, Korea and identified by Professor Jae-Mo Sung (Department of Agricultural Biology of Kangwon National University). The dried cultured fruit bodies were extracted with distilled deionized water (DDW) for 6 hrs at 60℃. The water extracts were concentrated with a vacuum evaporator at 70℃ then lyophilized. Cordyceps militaris extract (CME) was included the cordycepin (3'-deoxyadenosine) 1.3 ng/mg which was purchased from Sigma (St. Louis. MO, USA).
RAW264.7 mouse macrophage cells (American Type Culture Collection) were maintained in Dulbecco's Modified Eagle's Medium (DMEM), supplemented with 10% heat-inactivated fetal bovine serum (FBS; Hyclone, USA), 100 U/ml of penicillin, and 100 g/ml of streptomycin (Hyclone, USA) at 37℃ in a 5% CO2 incubator. Primary macrophages were collected from the peritoneal cavities of mice (8-week-old male ICR mice) after an intraperitoneal (i.p.) injection of 3 ml of 3% thioglycolate broth (Sigma, St. Louis, MO) 4 days before harvesting. The peritoneal macrophages were then washed with 1×PBS (Ca2+- and Mg2+-free) and plated with DMEM containing 10% FBS, 100 U/ml of penicillin, and 100 U/ml of streptomycin at 37℃ in a 5% CO2 incubator.
A commercially-available cell viability assay was employed to evaluate the cytotoxic effect of cordycepin using thiazolyl blue tetrazolium bromide (Sigma, St. Louis, USA). RAW264.7 cells (2×105 cells/well) were plated with various concentrations of CME in 96-well microtiter plates (Nunc, Roskilde, Denmark) and were then cultured overnight at 37℃ in a 5% CO2 incubator. Afterwards, 50 µl of MTT solution was added to each well, and the cells were then cultured for 4 hrs at 37℃ in a 5% CO2 incubator. 100 µL of solubilized solution were added to each well. The plate was allowed to stand overnight in the incubator after evaluation for complete solubilization of the purple formazan crystals and measurement of the optical density (OD) at 560 nm by a microplate reader (Molecular Devices corporation, Sunnyvale, USA).
The amount of nitrite and PGE2 produced by mouse macrophages was measured in cell culture supernatant. The cells were plated at a density of 1×106 cells in a flat-bottomed, 96-well plates with the 200 µl of culture medium. They were then treated with various concentrations (12.5~200 µg/ml) of CME in the absence of LPS (10 ng/ml) and incubated for overnight. NO production was determined according to the method reported by Stuehr and Nathan (21). The amount of nitrite was measured using the Griess reagent (stock-I: 0.2% N-(1-naphthyl) ethylenediamine-HCl, stock-II: 2% sulfanilamide in 5% H2PO4) and the amount of PGE2 produced was measured using an enzyme-linked immunosorbent assay (ELISA) kit (R&D system, Minneapolis, MI, USA), according to the manufacturer's instructions.
The amount of TNF-α, IL-1β, and IL-6 in the cell culture supernatant were measured using an ELISA kit (eBioscience, San Diego, CA, USA). RAW 264.7 cells were cultured in DMEM with 10% FBS in 12-well, flat-bottomed plates at a density of 5×105 cells/well. The cells were treated with various concentrations (12.5, 25, 50, 100, 200 µg/ml) of CME in the absence or presence of LPS (50 ng/ml) at 37℃ for 48 hrs in humidified air with 5% CO2. Subsequently, the culture supernatant was collected and assayed according to the manufacturer's instructions.
Total RNA was extracted from RAW264.7 cells using the RNeasy Mini kit (QIAGEN, USA) in an RNase-free environment. RNA was quantified by reading the absorbance at 260 nm as previously described (14). The reverse transcription of 1 µg RNA was carried out using M-MLV reverse transcriptase (Promega, USA), oligo (dT) 16 primer, dNTP (0.5 µM), and 1 U RNase inhibitor. After incubation at 65℃ for 5 min and 37℃ for 60 min, M-MLV reverse transcriptase was inactivated by heating at 70℃ for 15 min. The polymerase chain reaction (PCR) was performed in 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, and 2.5 mM dNTPs with 5 units of Taq DNA polymerase and 10 pM of each primer set for iNOS, PGE2, IL-1β, IL-6, and PGE2. The cDNA was amplified by 35 cycles of denaturing at 94℃ for 45 s, annealing at 62℃ for 45 s, and extension at 72℃ for 1 min. Final extension was performed at 72℃ for 5 min. The PCR products were electrophoresed on a 1.5% agarose gels and stained with ethidium bromide. The primers used were 5' AGCTCCTCCCA GGACCACAC 3' (forward) and 5' ACGCTGAGTACCTCATTGGC 3' (reverse) for iNOS, 5' CAGGATGAGACATGACACC 3' (forward) and 5' CTCTGCAGACTCAAACTCCAC 3' (reverse) for IL-1β, 5' GTACTCCAGAAGACCAGAGG 3' (forward) and 5' TGCTGGTGACAACCACGGCC 3' (reverse) for IL-6, 5' TTGACCTCAGCGCTGAGTTG 3' (forward) and 5' CCTGTAGCCCACGTCGTAGC 3' (reverse) for TNF-α, 5' AAGAAGAAAGTTCATTCCTGATCCC 3' (forward) and 5' TGACTGTGGGAGGATACATCTCTC 3' (reverse) for COX-2, and 5' GTGGGCCGCCCTAGGACCAG 3' (forward) and 5' GGAGGAAGAGGATGCGGCAGT 3' (reverse) for β-actin. β-actin was used as an internal control.
After cultured cells were collected and washed twice with cold PBS, they were resuspended in hypotonic buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.2 mM PMSF, 0.5 mM DTT, 10 µg/ml aportinin). After 15 min incubation on ice, the cells were lysed by the addition of 0.1% NP-40 and vigorous vortexing for 1 min. The nuclei were pelleted by centrifugation at 12,000×g for 1 min at 4℃ and resuspended in high salt buffer (20 mM HEPES, pH 7.9, 25% glycerol, 400 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 1 mM NaF, 1 mM sodium orthovanadate). The supernatant fluid was stored in aliquots at -70℃.
RAW 264.7 cells were washed with phosphate-buffered saline (PBS) and lysed by lysis buffer (1% SDS, 1.0 mM sodium vanadate, 10 mM Tris-Cl buffer, pH 7.4) for 5 min. 20 µg of protein from the cell lysates was applied to 8~12% SDS-polyacrylamide gels and then transferred to nitrocellulose membranes. The membranes were blocked with 5% skim milk in PBST solution for 1 hr. They were then incubated with anti-IL-1β, anti-IL-6, anti-TNF-α, anti-i-NOS, anti-COX-2, and anti-NF-κB monoclonal antibodies (Santa Cruz Biotechnology Inc., Santa Cruz, California, USA) for 2 hrs and washed 3 times with PBST. After incubation with an alkaline phosphatase-labeled secondary antibody (Abcam, Cambridge, USA) for 2 hrs, the bands were visualized using a Western Blot Kit with alkaline phosphatase substrate (Vector, Burlingame, USA).
RAW 264.7 cells (1×106 cells/ml) were cultured in Petridishes. The cells were treated with various concentrations (12.5, 50, 200 µg/ml) of CME in the absence or presence of LPS (100 ng/ml). the dishes were incubated at 37℃ for 24 hrs in humidified 5% CO2 incubator under standard conditions. The cells were then washed with PBS. The washed cells were blocked with staining buffer containing 10% normal mouse serum (NMS) for 20 min on ice. The blocked cells were incubated with co-stimulatory molecules such as ICAM-1, B7-1 and B7-2 antibody (BD Biosciences, San Jose, USA) for 20 min on ice. The incubated cells were washed three times with staining buffer and then fixed by 1% paraformaldehyde in PBS. The fixed cells were measured by flow cytometry (Beckman coulter, Brea, USA).
Data are expressed as mean±standard deviation. Statistical significance between the groups was determined by a paired t-test and one-way ANOVA for repeated measures. Results of p<0.05 were considered statistically significant. Data were assessed using SPSS (version 15.0, SPSS Inc., Chicago, Illinois).
To analyze the potential proinflammatory properties of CME, we used murine macrophages, which produces NO and PGE2. Nitric oxide is known to represent toxic and proinflammatory mediators in acute and chronic inflammatory diseases as well as in normal defense reactions. In the current study, macrophages did not release NO in response to the medium alone; LPS (10 ng/ml) was used as a positive control for macrophage activation (Fig. 1A and B). When various concentrations of CME (12.5, 25, 50, 100, 200 µg/ml) were added to the culture media at the time of cell stimulation (18 hrs), NO production increased in a CME concentration-dependent manner (Fig. 1A and B). Further, CME was also found to dose-dependently increase PGE2 production (Fig. 1C and D). The potential cytoxicity of CME was evaluated by MTT assay after incubating cells for 24 hrs in the absence or presence of LPS. Results revealed that viability was unaffected at the concentrations used to enhance NO and PGE2 (data not shown). Thus, the immune enhancement was not attributable to a cytotoxic effect.
RT-PCR and western blot analyses were performed to determine whether CME had a direct effect on proinflammatory mediators (NO and PGE2) related to modulation of the expression of iNOS and COX-2. As shown in Fig. 2A, iNOS and COX-2 protein expression was markedly induced in macrophage cells after treatment both with CME and LPS. This induction was increased by CME treatment in a dose-dependently manner. Furthermore, RT-PCR analysis revealed that the expression of the COX-2 gene was correlated with their protein levels (Fig. 2B).
To determine whether CME had a direct effect on cytokine production, IL-1β, IL-6, and TNF-α secretion were measured in the macrophage cell line using cytokine ELISA kits. IL-1β, IL-6, and TNF-α, are the major proinflammatory cytokines produced by monocytes and macrophages. As shown in Fig. 3, CME dose-dependently increased cytokine production. Interestingly, CME also strongly increased IL-1β and IL-6 production.
To further investigate the important role of CME on immune response, inducing RAW 264.7 cells for 24 hours with LPS or CME resulted in the production of proinflammatory cytokines. As shown in Fig. 4A, CME concentration-dependently induced the expression of IL-1β, IL-6, and TNF-α protein and mRNA (Fig. 4B).
Further to evaluate whether CME influences the phenotypic maturation of macrophages were cultured with CME or LPS for 24 h and then analyzed for any surface expression of ICAM-1, B7-1, and B7-2 molecules. As shown in Fig. 5, CME similarly to LPS, induced dramatic up-regulation of ICAM-1 and B7-1/-2 molecules.
It has been shown that NF-κB activation is a critical factor for the expression of various cytokines, iNOS, and COX-2 in macrophages in response to LPS. In an unstimulated cell, NF-κB resides in the cytoplasm as an inactive NF-κB/IκB complex. Under stimuli, IκBα is phosphorylated and subsequently degraded, allowing NF-κB to translocate into the nucleus (13). To investigate whether CME could affect nuclear translocation of NF-κB, a western blot analysis for NF-κB p65 was carried out with cell lysate in macrophages (Fig. 6). The amount of NF-κB p65 markedly increased upon exposure to LPS alone; CME also increased NF-κB p65.
The data presented in this paper indicated that CME can exert significant immunomodulatory effects on macrophage-mediated immune responses. The present study demonstrated that CME is an effective activator of NO generation, cytokine (IL-1β, IL-6, and TNF-α) expression, PGE2 secretion, and co-stimulatory molecules in macrophages. Inflammation is a complex process involving numerous mediators of cellular and plasma origins. Activated macrophages fuse their lysosomes more efficiently to phagosomes, exposing intracellular or recently ingested extracellular microbes to a variety of microbiocidal lysosomal enzymes. Activated macrophages also produce oxygen radicals and NO, both of which have potent antimicrobial activity. Furthermore, they synthesize anti-microbial peptides and proteases that can be released to attract extracellular parasites. Additional changes in activated macrophages assist in amplifying the immune response.
Nitric oxide has been known to play a critical role in prevention against microbial infection and cancer occurrence (23,24). In addition, NO may be involved with regulation of apoptosis (25) and progression of autoimmune disease by altering the differentiation of the helper T cell phenotype (26). The biological activity of NO has become more complicated since NO production was reported to suppress the functional activity of T lymphocytes and antigen presenting cells (APCs) (27). In this paper, we have demonstrated that CME induced an increase of NO production in vitro (Fig. 1 and 2).
Prostaglandins, which are produced by COX-2, have been implicated as important mediators in endotoxemia and inflammatory conditions (28). For example, PGE2, like NO, is a pleiotropic mediator produced at inflammatory sites by COX-2 that gives rise to pain, swelling and stiffness (29). Here, we have demonstrated that CME significantly increases the gene expression of COX-2 and PGE2 production in macrophage cells (Fig. 1 and 2).
Cytokines are local protein mediators and are involved in almost all important biological processes including cell growth and activation, inflammation, immunity, and differentiation. In this study, CME dose-dependently enhanced IL-1β, IL-6 and TNF-α production in RAW 264.7 cells (Fig. 3 and 4).
The expression of cytokines requires the activation of NF-κB, a nuclear transcription factor, which regulates the expression of various genes, including IL-1β, i-NOS, and COX-2 all of which play critical roles in apoptosis and autoimmune disease. Nuclear factor-κB requires the phosphorylation of IκB and then targets IκB for ubiquitination and degradation. As shown in Fig. 6, CME increased proinflammatory mediators via NF-κB activation in RAW 264.7 cells.
The intracellular adhesion molecules (ICAMs), ICAM-1, ICAM-2 and ICAM-3, are cell surface ligands for leukocyte integrins. They are crucial in the binding of lymphocytes and other leukocytes to certain cells including APCs. In the current study, CME induced the ICAM-1 molecules in macrophages (Fig. 5A). The B7 family plays an important role as a co-stimulatory factor in APCs. Treatment with CME had another major effect on co-stimulatory molecules B7-1/-2 by strongly upregulating the surface levels of B7-1/-2 molecules in macrophage cells (Fig. 5B, C). All these molecules have been described to be of major importance in APC function (30,31).
In summary, the finding of the present study suggest that CME isolated from the fruiting bodies of Cordyceps militaris is a potent activator of NO, PGE2, and proinflammation production in macrophages, and that it acts at the level of transcription. Moreover, these inductions of the immune reaction by CME were associated with NF-κB activation. In conclusion, CME has immunostimulatory effects on macrophages through an enhancement of the production of cytokines and other bioactive substances as well as through the increased expression of co-stimulatory and adhesion molecules. In conclusion, together with these facts of our finding suggest that CME might be useful as a therapeutic agent for the treatment of the immune deficiency diseases.
Figures and Tables
Figure 1
Effects of CME on NO and PGE2 production in macrophages. (A) RAW 264.7 cells and (B) peritoneal macrophages were treated with various concentrations (12.5, 25, 50, 100, 200 µg/ml) of CME or LPS (10 ng/ml), then incubated overnight. Nitrite levels in culture media were determined using Griess reagent and were presumed to reflect NO levels. (C) RAW 264.7 cells and (D) peritoneal macrophages were treated with 12.5~200 µg/ml of CME for 48 hrs. Levels of PGE2 in culture media were quantified using EIA kits. Data are presented as means±S.D. of three independent experiments. †p<0.01 vs. cells only; *p<0.05, **p<0.01 vs. cells only.

Figure 2
Effects of CME on the expression of iNOS and COX-2 protein and mRNA in macrophages. (A) RAW 264.7 cells were treated with various concentrations (12.5, 25, 50, 100, 200 µg/ml) of CME or LPS (100 ng/ml) for 24 hrs. Total cellular proteins (20 µg) were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and detected with specific antibodies, as described in Materials and Methods. (B) Cells were treated as described above. Total RNA was prepared for the RT-PCR analysis of the expression of iNOS and COX-2 in macrophages treated with various concentrations (12.5, 25, 50, 100, 200 µg/ml) of CME or LPS (100 ng/ml) for overnight. iNOS-specific sequences and COX-2-specific sequences were detected by agarose gel electrophoresis, as described in Materials and Methods. PCR for β-actin was performed to verify that the initial cDNA contents of the samples were similar. This was repeated in triplicate and similar results were obtained in all three.
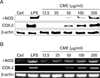
Figure 3
Effect of CME on cytokine production in RAW 264.7 cells. Effects of CME on IL-1β (A), IL-6 (B), and TNF-α (C) production in macrophages. Cultures were treated with various concentrations of CME (12.5, 25, 50, 100, 200 µg/ml). The supernatants were collected, and the extracellular levels of cytokines were measured in culture media using cytokine ELISA kits. The results are reported as mean±S.D. of three independent experiments. †p<0.01 vs. cells only; *p<0.05, **p<0.01 vs. cells only.

Figure 4
Effects of CME on the expression of pro-inflammatory cytokines proteins and mRNA in macrophages. (A) RAW 264. 7 cells were treated with various concentrations (12.5, 25, 50, 100, 200 µg/ml) of CME or LPS (100 ng/ml) for 24 hrs. Total cellular proteins (20 µg) were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and detected with specific antibodies, as described in the Materials and Methods. (B) Cells were treated as described above. Total RNA was prepared for the RT-PCR analysis of the expression of IL-1β, IL-6, and TNF-α in macrophages treated with various concentrations (12.5, 25, 50, 100, 200 µg/ml) of CME or LPS (100 ng/ml) for overnight. IL-1β-specific sequences, IL-6-specific sequences, and TNF-α-specific sequences were detected by agarose gel electrophoresis, as described in Materials and Methods. PCR for β-actin was performed to verify that the initial cDNA contents of the samples were similar. The experiment was repeated in triplicate and similar results were obtained in all three.
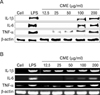
Figure 5
Effects of CME on the expression of co-stimulatory molecules in RAW 264.7 cells. Cells were cultured with various concentrations of CME (12.5, 50, 200 µg/ml) or LPS (100 ng/ml) for 24 hrs. Surface ICAM-1 (A), B7-1 (B), and B7-2 (C) molecules were labeled with either anti-ICAM-1 or anti-B7-1/-2.

Figure 6
Effects of CME on NF-κB activation in RAW 264.7 cells. Cells were incubated overnight with various concentrations of CME (12.5~200 µg/ml) in the absence or presence of LPS (100 ng/ml). Protein (20 µg) from each sample was resolved in 12% SDS-PAGE and then analyzed by western blotting. β-actin was used as a control.

References
1. Gai G, Jin S, Wang B, Li Y, Li C. The efficacy of Cordyceps militaris capsules in treatment of chronic bronchitis in comparison with Jinshuibao capsules. Chin J New Drugs. 2004. 13:169–171.
2. Choi SB, Park CH, Choi MK, Jun DW, Park S. Improvement of insulin resistance and insulin secretion by water extracts of cordyceps militaris, phellinus linteus, and paecilomyces tenuipes in 90% pancreatectomized rats. Biosci Biotechnol Biochem. 2004. 68:2257–2264.

3. Yun Y, Han S, Lee S, Ko S, Lee C, Ha N, Kim K. Anti-diabetic effects of CCCA, CMESS, and cordycepin from Cordyceps militaris and the immune responses in streptozotocin-induced diabetic mice. Nat Prod Sci. 2003. 9:291–298.
4. Yu R, Wang L, Zhang H, Zhou C, Zhao Y. Isolation, purification and identification of polysaccharides from cultured Cordyceps militaris. Fitoterapia. 2004. 75:662–666.

5. Cho MA, Lee DS, Kim MJ, Sung JM, Ham SS. Antimutagenicity and cytotoxicity of cordycepin isolated from Cordyceps rnilitaris. Food Sci Biotechnol. 2003. 12:472–475.
6. Won SY, Park EH. Anti-inflammatory and related pharmacological activities of cultured mycelia and fruiting bodies of Cordyceps militaris. J Ethnopharmacol. 2005. 96:555–561.

7. Chen C, Luo S, Sun YJ, Zhang CK. Study on antioxidant activity of three Cordyceps sp. Chin J Biochem Pharm. 2004. 25:212–214.
8. Ross JA, Auger MJ, Burke B, Lewis CE. Burke B, Lewis CE, editors. The biology of the macrophage. The macrophage. 2002. 2nd ed. Oxford, UK: Oxford Medical Publications;1–72.
9. Kinne RW, Bräuer R, Stuhlmuller B, Palombo-Kinne E, Burmester GR. Macrophages in rheumatoid arthritis. Arthritis Res. 2000. 2:189–202.
10. Nathan C. Inducible nitric oxide synthase: what difference does it make? J Clin Invest. 1997. 100:2417–2423.

12. Vila-del Sol V, Fresno M. Involvement of TNF and NF-kappa B in the transcriptional control of cyclooxygenase-2 expression by IFN-gamma in macrophages. J Immunol. 2005. 174:2825–2833.

13. Isomäki P, Punnonen J. Pro- and anti-inflammatory cytokines in rheumatoid arthritis. Ann Med. 1997. 29:499–507.
14. Libby P, Ridker PM, Maseri A. Inflammation and atherosclerosis. Circulation. 2002. 105:1135–1143.

15. Tilg H, Wilmer A, Vogel W, Herold M, Nölchen B, Judmaier G, Huber C. Serum levels of cytokines in chronic liver diseases. Gastroenterology. 1992. 103:264–274.

16. Coker RK, Laurent GJ. Pulmonary fibrosis: cytokines in the balance. Eur Respir J. 1998. 11:1218–1221.

17. Lawrence T, Gilroy DW, Colville-Nash PR, Willoughby DA. Possible new role for NF-kappaB in the resolution of inflammation. Nat Med. 2001. 7:1291–1297.

18. Riehemann K, Behnke B, Schulze-Osthoff K. Plant extracts from stinging nettle (Urtica dioica), an antirheumatic remedy, inhibit the proinflammatory transcription factor NF-kappaB. FEBS Lett. 1999. 442:89–94.

19. Renard P, Raes M. The proinflammatory transcription factor NFkappaB: a potential target for novel therapeutical strategies. Cell Biol Toxicol. 1999. 15:341–344.
20. Makarov SS. NF-kappaB as a therapeutic target in chronic inflammation: recent advances. Mol Med Today. 2000. 6:441–448.
21. Stuehr DJ, Nathan CF. Nitric oxide. A macrophage product responsible for cytostasis and respiratory inhibition in tumor target cells. J Exp Med. 1989. 169:1543–1555.

22. Majumder N, Dey R, Mathur RK, Datta S, Maitra M, Ghosh S, Saha B, Majumdar S. An unusual pro-inflammatory role of interleukin-10 induced by arabinosylated lipoarabinomannan in murine peritoneal macrophages. Glycoconj J. 2006. 23:675–686.

23. MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997. 15:323–350.

24. Underhill DM, Ozinsky A. Phagocytosis of microbes: complexity in action. Annu Rev Immunol. 2002. 20:825–852.

25. Li J, Billiar TR, Talanian RV, Kim YM. Nitric oxide reversibly inhibits seven members of the caspase family via S-nitrosylation. Biochem Biophys Res Commun. 1997. 240:419–424.

26. Kolb H, Kolb-Bachofen V. Nitric oxide in autoimmune disease: cytotoxic or regulatory mediator? Immunol Today. 1998. 19:556–561.

27. Paterson RR. Cordyceps: a traditional Chinese medicine and another fungal therapeutic biofactory? Phytochemistry. 2008. 69:1469–1495.

28. Ahmad N, Chen LC, Gordon MA, Laskin JD, Laskin DL. Regulation of cyclooxygenase-2 by nitric oxide in activated hepatic macrophages during acute endotoxemia. J Leukoc Biol. 2002. 71:1005–1011.
29. Seibert K, Zhang Y, Leahy K, Hauser S, Masferrer J, Perkins W, Lee L, Isakson P. Pharmacological and biochemical demonstration of the role of cyclooxygenase 2 in inflammation and pain. Proc Natl Acad Sci USA. 1994. 91:12013–12017.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download