Abstract
In order to perform large-scale genetic studies of Kawasaki disease (KD) in Korea, the Korean Kawasaki Disease Genetics Consortium (KKDGC) was formed in 2008 with 10 hospitals. Since the establishment of KKDGC, there has been a collection of clinical data from a total of 1198 patients, and approximately 5 mL of blood samples per patient (for genomic deoxyribonucleic acid and plasma isolation), using a standard clinical data collection form and a nation-wide networking system for blood sample pick-up. In the clinical risk factor analysis using the collected clinical data of 478 KD patients, it was found that incomplete KD type, intravenous immunoglobulin (IVIG) non-responsiveness, and long febrile days are major risk factors for coronary artery lesions development, whereas low serum albumin concentration is an independent risk factor for IVIG non-responsiveness. In addition, we identified a KD susceptibility locus at 1p31, a coronary artery aneurysm locus (KCNN2 gene), and the causal variant in the C-reactive protein (CRP) promoter region, as determining the increased CRP levels in KD patients, by means of genome-wide association studies. Currently, this consortium is continually collecting more clinical data and genomic samples to identify the clinical and genetic risk factors via a single nucleotide polymorphism chip and exome sequencing, as well as collaborating with several international KD genetics teams. The consortium-based approach for genetic studies of KD in Korea will be a very effective way to understand the unknown etiology and causal mechanism of KD, which may be affected by multiple genes and environmental factors.
Kawasaki disease (KD) is an acute self-limiting form of vasculitis that afflicts infants and children and manifests itself as fever and signs of mucocutaneous inflammation. KD is characterized by prolonged fever, bilateral conjunctival infection, erythema of the oral mucosa, lips, and tongue, polymorphous rash, erythema of the palms and soles, and cervical lymphadenopathy.1) Damage to the coronary arteries occurs in 15-25% of untreated individuals, and this has made KD the leading cause of pediatric acquired heart disease in developed countries.2) Treatment with high-dose intravenous immunoglobulin (IVIG) markedly reduces fever duration, systemic inflammation, and coronary artery lesion (CAL) in children with KD, but about 10% of patients are unresponsive to IVIG and demonstrate persistent or recurrent fever after initial IVIG treatment.2) The average annual incidence of KD in Korea is 134.4 per 100000 children <5 years of age in 2011, which is the second highest incidence of KD worldwide, following its incidence in Japan.3) To date, the etiology of KD is still largely unknown. Thus, no diagnostic test or prevention is available. In order to facilitate the genetic studies of KD, the Korean Kawasaki Disease Genetics Consortium (KKDGC) was formed in 2008 with 10 hospitals. In this review paper, the overall research interests of the KKDGC are introduced, as well as its standardized forms and procedures.
In order to collect a large number of the KD patients' clinical data and genomic deoxyribonucleic acid (DNA) samples for genetic studies of KD in Korea, the first KKDGC was formed in 2008 with 10 hospitals and 1 company (Seoul Clinical Laboratories, SCL). The consortium initially had set a goal to collect a total of 500 KD samples and achieved this goal in less than 2 years (May 2008 to February 2010) by collecting a total of 517 KD samples. The collected clinical data and genomic DNA samples were used for clinical data analysis and genetic studies of KD. However, it was realized that 500 KD case samples were not sufficient for large-scale genetic studies of KD. Thus, the second consortium was started in 2012 (April 2012 to September 2014). Through the first and second consortia, a total of 1198 KD case samples (genomic DNA and plasma samples) and clinical data were collected (Fig. 1). The collection of clinical data and genomic DNA samples is still ongoing by way of the second KKDGC.
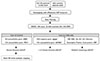
The KD patients were recruited from 13 tertiary academic hospitals in Korea that are currently participating in the KKDGC. All the KD patients were diagnosed by pediatricians, in accordance with the diagnostic criteria of the American Heart Association.2) At each participating hospital, approximately 5 mL of blood from a KD patient was sampled in an acid citrate dextrose or ethylenediamine tetraacetic acid tube after explanation and approval with the informed consents form. The collected patients' blood samples were used for genomic DNA preparation or establishment of Epstein-Barr Virus-transformed B cell lines at SCL (Fig. 2). Each patient's sample and clinical data were recorded by a new sample identification (ID) system that uses a two-digit disease symbol (KD), two-digit hospital ID, and a 3 digit-serial number (e.g., KD-AS-001). Pediatricians at each participating hospital collected all clinical data of the KD patients, including detailed clinical signs and treatment response. Finally, all the patients' clinical data and genomic DNA samples were deposited at Asan Medical Center at Seoul.
In order to characterize the clinical features of Korean children with KD prior to genetic studies, we constructed a standard clinical data collection form of KD with a total of 41 variables. The clinical data collection sheet is composed of: 1) patient's personal information, 2) clinical signs and symptoms, 3) echocardiogram findings, 4) treatment responses, 5) family history and recurrence, and 6) clinical lab data (Fig. 3). For genetic association studies of the KD subgroup, the clinical data were used to categorize the patient subgroups into a KD type (complete vs. incomplete KD), KD patients with or without CALs (normal vs. CAL), or IVIG responsiveness (responder vs. non-responder).
Using the clinical data of 478 KD patients collected during the first consortium, there was an investigation of the clinical features and risk factors associated with CAL development and IVIG nonresponsiveness in Korean children with KD. We found that incomplete KD type, IVIG nonresponse, fever duration of 7 days or longer, and CC/AC genotypes of the rs7604693 single nucleotide polymorphism (SNP) in the PELI1 gene, were significantly associated with the development of CALs, with odds ratios (ORs) ranging from 2.06 to 3.04.4) This study also found that a serum albumin level of 3.6 g/dL or lower was significantly associated with nonresponsiveness to IVIG (OR=2.76; p=0.006).4)
To examine the mechanism of IVIG-resistance in KD patients, we initially performed cell proliferation assays after in vitro immunoglobulin treatment in various cell lines, including KD patient-derived B cell lines (either IVIG responder or IVIG non-responder patient). Both the KD patient-derived B cell lines equally exhibited dose-dependent cell death by in vitro immunoglobulin treatment. However, no difference in cell death was observed in either B cell lines derived from the responder KD patient or non-responder KD patient (Fig. 4A). Other cell lines also underwent cell death by in vitro immunoglobulin treatment (Fig. 4B and C). This result suggests that the B cell may not play a role for IVIG responsiveness in KD patients. In addition, to understand the mechanisms of IVIG resistance in KD patients, we examined the gene expression profiles of B cell lines derived from IVIG responsive (n=2) or IVIG nonresponsive KD patients (n=3). We observed 34 upregulated (>2-fold) and 14 downregulated (<2 fold) genes after in vitro immunoglobulin treatment in B cells.5) Specifically, there was significant upregulation of gene expression in immune-related genes and downregulation in genes involved in cell-cycle after in vitro immunoglobulin treatment in B cell lines. However, there was no difference in the pattern of gene expression between IVIG responders and IVIG non-responders. This result suggests that the resistance of IVIG treatment in KD patients is not mediated by the change of quantitative gene expression in specific genes after IVIG treatment. Furthermore, in vitro immunoglobulin treatment in B cell lines did not increase the expression of the inhibitory FCGR2B gene. Therefore, this data suggest that the therapeutic effect of IVIG treatment in KD patients is not mediated by either inhibitory Fc receptor (FCGR2B) on B cells or B cell-mediated immune responses.
We performed a Genome-Wide Association Study (GWAS) using Affymetrix SNP array 6.0 in 186 KD patients and 600 healthy controls (Fig. 5). One SNP on chromosome 1p31 (rs527409) and a PELI1 locus on chromosome 2p13.3 (rs7604693) was associated with KD susceptibility and CAL formation, respectively.6) A subgroup analysis was also performed to identify IVIG response genes and CAL susceptibility using case samples only. It was found that a SNP (rs17136627) in the KCNN2 gene was significantly associated with CAL formation, particularly with large aneurysms (diameter >5 mm) (OR=12.6, p=1.96×10-8).7) This result indicates that the KCNN2 gene can have an important role in the development of coronary artery aneurysms in KD. Furthermore, we also found a significant association of a CRP promoter SNP (rs12068753) with a high CRP level in KD (beta=3.97, p=1.11×10-13) via the genetic association analysis of inflammatory biomarkers in KD patients.8) In addition, we collaborated with international KD genetics groups, including International KD Genetics Consortium, Taiwan KD Genetics Consortium, and Japan KD Genetics Consortium. In particular, we contributed to the studies validating the FCGR2A association with KD, performed by the international KD Genetics Consortium,9) and replication of B lymphoid tyrosine kinase association with KD that was carried out by Taiwan KD Genetics Consortium.10) In addition, the meta-analysis of Asian KD GWAS data is under way with the Japanese and Taiwanese groups.
Currently, we are re-analyzing the clinical data of all collected KD patients (n=~1200) to evaluate the effect of age, gender, family history status, recurrence status, and KD types, on clinical features of KD. We also plan to study the interaction of clinical risk factors and genetic risk factors in KD. In order to identify new KD susceptibility and subphenotype loci, we are performing another GWAS using an Illumina Human Omni1 SNP chip with approximately 300 KD cases, concerning 16 cases with family history, 46 cases with recurrence, 119 cases with IVIG non-responsiveness, and 52 cases with CALs (diameter >5 mm). Multiple subsets of KD cases will be very useful to detect the loci associated with the subphenotypes of KD in GWAS data analysis. In the near future, we also plan to adopt the exome sequencing approach for KD genetic studies. As demonstrated above, consortium-based genetic studies are an effective way to identify the clinical risk factors and genetic risk factors of KD that may be affected by multiple genes and environmental factors. Furthermore, the KKDGC will facilitate and contribute to the understanding of pathophysiological mechanisms of KD and the development of novel diagnosis, as well as treatment and prevention strategies.
Figures and Tables
Fig. 1
Participating hospitals in KKDGC and the numbers of sample collected. A total of 517 and 681 KD case samples were collected during the first (May 2008 to February 2010) and second (April 2012 to September 2014) KKDGC, respectively. KKDGC: Korean Kawasaki Disease Genetics Consortium, KD: Kawasaki disease.

Fig. 2
Work process of KKDGC to collect the clinical data and genomic DNA samples. Each patient's blood sample and clinical data were collected using standard protocol. KKDGC: Korean Kawasaki Disease Genetics Consortium, DNA: deoxyribonucleic acid, AMC: Asan Medical Center, SCL: Seoul Clinical Laboratories, DB: database, IRB: institutional review board, ACD: acid citrate dextrose, EDTA: ethylenediamine tetraacetic acid, RT: room temperature, EBV: Epstein-Barr virus.

Fig. 3
Clinical data collection sheet for KD. A total of 41 clinical variables were collected per patient. KD: Kawasaki disease, ID: identification, IVIG: intravenous immunoglobulin, CRP: C-reactive protein, ESR: erythrocyte sedimentation rate, Hb: hemoglobulin, AST: aspartate aminotransferase, ALT: alanine aminotransferase, IU: international unit, L: liter.

Fig. 4
Cell proliferation assay after in vitro IG treatment in KD patient-derived B cell lines (A), macrophage-like cell line (U937) (B) and neutrophil-like cell (HL60) (C). Each cell was treated with various concentrations of IG for 48 hr and cell proliferation assay was performed using commercial kit. IG: immunoglobulin, KD: Kawasaki disease.

Fig. 5
Overview of GWAS performed by KKDGC. The results of GWAS were published.6)7)8) GWAS: genome-wide association study, KKDGC: Korean Kawasaki Disease Genetics Consortium, KD; Kawasaki disease, M: male, F: female, SNP: single nucleotide polymorphism, CAL: coronary artery lesion, IVIG: intravenous immunoglobulin, CRP: C-reactive protein, WBC: white blood cell, Hb: hemoglobulin, AST: aspartate aminotransferase, ALT: alanine aminotransferase.
Acknowledgments
We thank all the patients with KD and their families for participating in this study. This study was supported by grants (A010384, A111517) from the Korea Ministry of Health and Welfare, grant (2014-ER740200) from the Korea Centers for Disease Control and Prevention and grant (2011-419 & 2012-419) from the Asan Institute for Life Sciences, Seoul, Korea.
Korean Kawasaki Disease Genetics Consortium: Jeong-Jin Yoo*, In-Sook Park, Kwi-Joo Kim (Asan Medical Center, Seoul); Myung Ki Han* (Asan Medical Center, Gangneung); Jong-Keuk Lee*, Jae-Jung Kim, Young-Mi Park (Asan Institute for Life Sciences, Seoul); Sin Weon Yun* (Chung-Ang University Hospital, Seoul); Hong-Ryang Kil* (Chungnam National University Hospital, Daejeon); Young Mi Hong*, Saejung Sohn (Ewha Womans University Hospital, Seoul); Gi Young Jang*, Kee-Soo Ha, Hyo-Kyoung Nam, Jung-Hye Byeon (Korea University Hospital, Seoul); Kyung Lim Yoon* (Kyung Hee University Hospital, Seoul); Min Seob Song* (Inje University Paik Hospital, Busan); Hyoung Doo Lee* (Pusan National University Hospital, Busan); Jae-Moo Lee, Jong-Duk Kim (Seoul Clinical Laboratories, Seoul); Gi Beom Kim* (Seoul National University Children's Hospital, Seoul); Kyung-Yil Lee*, Jung-Woo Rhim (The Catholic University St. Mary's Hospital, Daejeon); Dong Soo Kim* (Yonsei University Severance Children's Hospital, Seoul). *Primary investigator at each institute.
References
1. Burns JC, Glodé MP. Kawasaki syndrome. Lancet. 2004; 364:533–544.
2. Newburger JW, Takahashi M, Gerber MA, et al. Diagnosis, treatment, and long-term management of Kawasaki disease: a statement for health professionals from the Committee on Rheumatic Fever, Endocarditis and Kawasaki Disease, Council on Cardiovascular Disease in the Young, American Heart Association. Circulation. 2004; 110:2747–2771.
3. Kim GB, Han JW, Park YW, et al. Epidemiologic features of Kawasaki disease in South Korea: data from nationwide survey, 2009-2011. Pediatr Infect Dis J. 2014; 33:24–27.
4. Kim JJ, Hong YM, Yun SW, et al. Assessment of risk factors for Korean children with Kawasaki disease. Pediatr Cardiol. 2012; 33:513–520.
5. Kim HE, Kim JJ, Han MK, et al. Variations in the number of CCL3L1 gene copies and Kawasaki disease in Korean children. Pediatr Cardiol. 2012; 33:1259–1263.
6. Kim JJ, Hong YM, Sohn S, et al. A genome-wide association analysis reveals 1p31 and 2p13.3 as susceptibility loci for Kawasaki disease. Hum Genet. 2011; 129:487–495.
7. Kim JJ, Park YM, Yoon D, et al. Identification of KCNN2 as a susceptibility locus for coronary artery aneurysms in Kawasaki disease using genome-wide association analysis. J Hum Genet. 2013; 58:521–525.
8. Kim JJ, Yun SW, Yu JJ, et al. Common variants in the CRP promoter are associated with a high C-reactive protein level in Kawasaki Disease. Pediatr Cardiol. 2015; 36:438–444.
9. Khor CC, Davila S, Breunis WB, et al. Genome-wide association study identifies FCGR2A as a susceptibility locus for Kawasaki disease. Nat Genet. 2011; 43:1241–1246.
10. Chang CJ, Kuo HC, Chang JS, et al. Replication and meta-analysis of GWAS identified susceptibility loci in Kawasaki disease confirm the importance of B lymphoid tyrosine kinase (BLK) in disease susceptibility. PLoS One. 2013; 8:e72037.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download