Abstract
Background
Methods
Results
Figures and Tables
Fig. 1
Comparison of the mitral inflow patterns in the diastolic stages between (A) the standard diet and (B) high-fat diet groups.
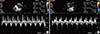
Fig. 2
Histomorphologic images stained with (A, B, ×200) H&E and (C, D, ×3,000; scale bar 1,000 nm) transmission electron microscopic images in the myocardia in the (A, C) standard diet and (B, D) high-fat diet groups.
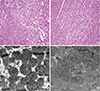
Fig. 3
The mRNA expression levels of a subset of oxidative phosphorylation (OXPHOS) subunits (A), PGC1α (B), antioxidant enzymes (C), Col1A1 (D), and SERCA2 (E) in the myocardia in the standard diet and high-fat diet groups. S, standard diet; H, high-fat diet; NDUFB3, NADH dehydrogenase 1 β subcomplex, 3; NDUFB5, NADH dehydrogenase 1 β subcomplex, 5; NDUFV1, NADH dehydrogenase flavoprotein 1; NDUFS1, NADH dehydrogenase Fe-S protein 1; SDHB, succinate dehydrogenase complex, subunit B; CYC1, cytochrome C1; SURF1, surfeit 1; PGC1α, peroxisome-proliferator-activated receptor γ, co-activator-1α; MnSOD, manganese-containing superoxide dismutase; Prdx1, peroxiredoxin 1; Prdx2, peroxiredoxin 2; Prdx5, peroxiredoxin 5; Col1A1, collagen, type I, α 1; SERCA2, sarcoendoplasmic reticulum calcium transport ATPase 2. aP<0.05; bP<0.01.

Fig. 4
Western blot results (A) and relative expression of pAKT (B), NDUFB5 (C), and PGC1α (D) in the myocardia in the standard diet and high-fat diet groups. S, standard diet; H, high-fat diet; pAKT, phospho-Akt; AKT, a serine/threonine kinase, protein kinase B; NDUFB5, NADH dehydrogenase 1-β subcomplex 5; PGC1α, peroxisome-proliferator-activated receptor γ, co-activator-1α; GAPDH, glyceraladehyde-3-phosphate dehydrogenase. aP<0.05.

Fig. 5
Mitochondrial DNA copy number (A), optical density of reactive oxygen species damage marker 8-OHdG (B), and ATP levels (C) in the myocardia in the standard diet and high-fat diet groups. S, standard diet; H, high-fat diet; 8-OHdG, 8-hydroxydeoxyguanosine; OD 450 nm, optical density of 8-OHdG as measured at 450 nm; ATP, adenosine triphosphate. aP<0.05.

Table 1
Real-Time Polymerase Chain Reaction Primer Sequences

NDUFB3, NADH dehydrogenase 1 β subcomplex, 3; NDUFB5, NADH dehydrogenase 1 β subcomplex, 5; NDUFV1, NADH dehydrogenase flavoprotein 1; NDUFS1, NADH dehydrogenase Fe-S protein 1; SDHB, succinate dehydrogenase complex, subunit B; CYC1, cytochrome C1; SURF1, surfeit 1; PGC1α, peroxisome-proliferator-activated receptor γ, co-activator-1α; MnSOD, manganese-containing superoxide dismutase; Prdx1, peroxiredoxin 1; Prdx2, peroxiredoxin 2; Prdx5, peroxiredoxin 5; COL1A1, collagen, type I, α 1; SERCA2, sarcoendoplasmic reticulum calcium transport ATPase 2; ND1, NADH dehydrogenase subunit 1; β-Globin, β-globin gene; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Table 2
Characteristics of Rats after 10 Weeks of a Standard Diet or a High-Fat Diet (n=6)

Table 3
Comparison of the Echocardiographic Parameters of Rats after 10 Weeks of a Standard Diet or a High-Fat Diet (n=6)

Values are expressed as mean±SD. Student t test was used to compare the parameters of the standard diet and high-fat diet groups.
LVPWS, left ventricular posterior wall at systole; LVIDS, left ventricular intraventricular dimension at systole; IVSS, interventricular septum at systole; LVPWD, left ventricular posterior wall at diastole; LVIDD, left ventricular intraventricular dimension at diastole; IVSD, interventricular septum at diastole; EF, ejection fraction; E, early wave in diastole; A, atrial wave in diastole; DT, deceleration time.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download