Abstract
Purpose
Tigecycline is one of the drugs used to treat multi-drug resistant Klebsiella pneumoniae (K. pneumoniae) infections, including complicated skin and soft tissue infections, complicated intra-abdominal infection, and community-acquired pneumonia in the Republic of Korea. However, since its commercial release, K. pneumoniae resistance against tigecycline has been reported, and there is a serious concern about the spread of tigecycline resistant bacteria.
Materials and Methods
In this study, we collected and analyzed 342 isolates from 23 hospitals in the Republic of Korea to determine the mechanisms of tigecycline susceptibility and their clonal types. The hospitals include several from each province in the Republic of Korea, except Jeju, an island province, and nonsusceptibility among the isolates was tested by the disk diffusion method. In our lab, susceptibility was checked again using the broth dilution method, and clonal types were determined using the multilocus sequence typing protocol. Real-time PCR was performed to measure the ramR mutation in the isolates nonsusceptible to tigecycline, which would suggest an increased expression of the AcrAB multidrug pump.
Results
Fifty-six K. pneumoniae isolates were found to be nonsusceptible, 16% of the 342 collected. Twenty-seven and nine isolates of the tigecycline nonsusceptible isolates had mutations in the ramR and rpsJ genes, respectively; while 18 nonsusceptible isolates harbored the tetA gene. Comparison of isolates with and without ramR mutation showed a significant statistical difference (p<0.05) for expression of AcrAB. Moreover, the most common clonal types, as observed in our study, appear to be ST11 and ST789.
Conclusion
Several dominate clonal types infer tigecycline resistance to K. pneumoniae, including ST11, ST768, ST15, ST23, ST48, and ST307. There does not seem to be a transferrable medium, such as plasmid, for the resistance yet, although mutation of the ramR gene may be a common event, accounting for 48% of the nonsusceptibility in this study.
Klebsiella pneumoniae (K. pneumoniae) is a member of the Enterobacteriaceae family, commonly found in the normal microbiota of human skin, and is also an important opportunistic pathogen that is known to cause pneumonia, urinary tract infection (UTI), cholecystitis, upper respiratory tract infection, and wound infection, among others. However, the emergence of multi-drug resistant (MDR) K. pneumoniae strains has posed a serious challenge in treating patients, especially in hospitals.1
Tigecycline is a minocycline derivative, which was among the first identified drugs from the class of glycylcyclines. It is a relatively new drug used for treating complicated skin and soft tissue infections (cSSTI), complicated intra-abdominal infection (cIAI), and community-acquired pneumonia in the Republic of Korea. It is also highly effective against many MDR strains of Enterobacteriaceae, including bacteria with the NDM gene.2 However, since its commercial release, there have been an increasing number of reports of resistance to tigecycline.34 Furthermore, there is a serious concern about the spread of tigecycline resistant bacteria. The known mechanisms of tigecycline resistance in Enterobacteriaceae to date are the tetA gene that encodes an efflux pump;56 a mutation in the S10 complex of the ribosome;7 and mutations in the ramR gene, which results in the overexpression of the AcrAB multi-drug pump.8
In this study, we collected 342 K. pneumoniae isolates from a network of tertiary hospitals that encompass all major provinces, except the island province of Jeju, in the Republic of Korea in 2012. Firstly, we screened for isolates that were nonsusceptible to tigecycline using the disk diffusion method. Next, we attempted to identify the antibiotic resistance mechanism and determine whether this attribute could spread to several other specific clonal types of K. pneumoniae.
Twenty-three tertiary hospitals in the Republic of Korea participated in this project, contributing seven to 15 unique clinical isolates from a total of 342 identified as K. pneumoniae. The isolates were screened for tigecycline nonsusceptibility using the disk diffusion method, including all isolates with the radius of the zone of 18 millimeters or less.9
Minimum inhibitory concentrations (MIC) of tigecycline and other antibiotics were evaluated for the clinical isolates by the broth dilution method (Sensititre GN2F, Thermo Scientific, Cleveland, OH, USA), and results were interpreted using CLSI 2012 breakpoints.
The presence of tet genes, such as tetA, B, C, D, and E, among the isolates was tested using PCR. In addition, we determined the sequences of ramR and rpsJ (ribosomal S10 protein), as described previously,7 for each of the clinical isolates and manually checked them for single nucleotide polymorphisms, insertions, and deletions that may have contributed to resistance to tigecycline. Furthermore, we used some of the most commonly known primers for the genes blaSHV, blaTEM, and blaCTX-M to identify markers for cephalosporin and carbapenem resistance among the samples.10
Multilocus sequence typing (MLST) was performed according to the published protocol (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html). The clonal types were determined by sequencing the amplified products and submitting them to the MLST website mentioned above. Simultaneously, we performed the eBURST analysis on the webserver http://eburst.mlst.net/ to identify the clonal complex.
After verifying the isolates with mutated ramR genes, we performed real-time quantitative PCR (qPCR) amplification to estimate the level of ramA expression. The primers were designed as previously described,7 and the SYBR green qPCR kit was used for quantification of expressed ramA. The isolates were normalized to isolate ID 23, which was shown not to have a mutation in the ramR gene by sequencing.
The disk diffusion method was used to screen the isolates for nonsusceptibility, zones with a radius larger than 18 millimeters. Most of the K. pneumoniae isolates were susceptible to tigecycline. However, 16% (56 out of 342) of the isolates demonstrated nonsusceptibility to tigecycline. Possibly because the isolates were selected using the disk diffusion method first, fifty-three isolates had an MIC of greater than 2, two isolates had an MIC of 0.5, and one isolate had an MIC of 2.
Nineteen nonsusceptible isolates for tigecycline were positive using PCR amplification for the tetA gene, while tetB, C, D, or E genes could not be detected. Upon sequencing and analysis of the ramR PCR amplification product, 27 mutations in the ramR gene were discovered; 12 of them co-existed with the tetA and six were exclusively associated with tetA. As shown in Table 1, eight of the 27 mutations encoded a stop codon, two of which were identical. Of the remaining 19, two mutations of the ST15 resulted in the missense mutation that resulted in a valine instead of alanine; two unrelated clonal types separately resulted in an isoleucine to threonine mutation in the same location. Two isolates of ST11 separately had ISKpn-18 inserts in different locations, as opposed to a previous report.7 The other mutations included deletion of a larger fragment, sequence insertion derived from another plasmid, insertion of a cytosine resulting in frameshift mutations such as; tyrosine to histidine, isoleucine to leucine, threonine to isoleucine, aspartic acid to histidine, lysine to glutamine, and a two-point mutation that resulted in a glycine to glutamic acid and aspartic acid to histidine separately.
Sequencing of the rpsJ PCR products yielded nine mutations with missense and single amino acid frameshift mutations. Out of the nine mutants, two co-existed with ramR mutations and four with the tetA gene. Isolates ST36 and 581 separately carried mutations in all three genes, namely ramR, rpsJ, and tetA.
Among the group of 56 nonsusceptible isolates for tigecycline, 37 and 36 were nonsusceptible to ceftazidime and ceftriaxone, respectively (Table 2). Seventeen CTX-M genes accounted for some of the resistance, although 55% of the resistance could not be attributed to a common extended spectrum of β-lactamase genes. Our samples also showed high cross resistance (39/56) with the fluoroquinolone ciprofloxacin, although we could not find any qnrA or qnrS genes by PCR amplification.
Thirty isolates from the 56 selected for nonsusceptibility to tigecycline were resistant to all three of the drugs, suggesting a cross-resistance between them.
A total of 31 clonal types were discovered as shown in Table 3, including four that had not been reported previously. The most frequent clonal type of K. pneumoniae that was nonsusceptible to tigecycline was ST11 (11 isolates), which had been reported to have tigecycline resistance previously described in Spain.11 The second most frequent clonal type with tigecycline resistance was ST789 (7 isolates), followed by 307 (4) and 23 and 48 (3 each), respectively.
The eBURST analysis showed that ST270 and ST1640 are single locus variants (SLVs) of ST11, while ST36 and ST268 are SLVs of ST504. ST793 and ST1637 were initially thought as newfound sequence type, but were found to be SLVs of ST23. Furthermore, ST280 and ST375 were SLVs of ST65. There were 10 other singletons, and the sequence types 1, 15, 20, 37, and 835 that were not related to any other sequence type were found in this study.
The real-time qPCR was performed using the gapA gene (F: CGAAACCGCTCGTAAACACA, R: AGGAAGCGTTGGAAACG ATG) as an internal control because the 16S rRNA and tonB genes provided unstable melting curves. For the external control, we used one of the isolates with a sequenced ramR gene carrying no mutations. The experiment was performed twice, and the statistical values were recorded (Table 4). Among the 27 isolates with ramR mutation, one isolate showed inconsistent real time PCR results, varying by 100-fold of the external control to 9000, thus it was not considered for further evaluation. t-test calculation of the two groups showed a p-value of 0.027, implying increased expression of ramA in the group with ramR mutations.
Reports on resistance of K. pneumoniae strains to tigecycline are ranked second in frequency to Acinetobacter baumanii.4 Moreover, the resistance of K. pneumoniae isolates to various antibiotics has risen sharply over the past decade.12 As one of the normal microbiota of the human gut and skin, K. pneumoniae pose one of the most important bacterial threats to immunocompromised and hospitalized patients. Tigecycline represents one of the last resort antibiotics for the treatment of MDR K. pneumoniae;2 meanwhile, several reports highlight the microbe's increased resistance to it.13
In our study, we identified several important tigecycline resistance factors and its spread in the Republic of Korea. First, the frequency of K. pneumoniae strains nonsusceptible to tigecycline is >16% across the nation, which may be quite high. There has been a report that tigecycline resistance occurs at a frequency of approximately 4×108 in the laboratory setting,14 and our study shows a similar occurrence in a clinical setting due to the selective pressure of antibiotics. Second, our study shows that the most common mechanism for tigecycline resistance in K. pneumoniae in the Republic of Korea seems due to independent mutations in ramR. In fact, ST11 and ST789, which were two of the most frequent isolates in our study, acquired independent and unrelated ramR mutations, implying that this gene is a genetic hotspot. Such genes accumulate spontaneous mutations as result of antibiotic pressure (over 16% of clinical cases) and may not require a medium, like plasmids, for dissemination. This highlights the effects of ramR gene in controlling the expression of the AcrAB multidrug pump, which is associated with nonsusceptibility to various drugs, including β-lactams, fluoroquinolones, and tigecycline, causing MDR in K. pneumoniae.15161718 We tested the isolates collected in this study (Table 1) for common carbapenemase and fluoroquinolone genes (blaVIM, blaIMP, blaGES, blaOXA-48, blaKPC, qnrS, and gnrA) and identified two and 39 isolates with nonsusceptibility to carbapenems and ciprofloxacin, respectively, although were unable to identify any associated genes through PCR amplification (data not shown). Barring two isolates, most of the isolates resistant to tigecycline were susceptible to carbapenems. Although mutation of ramR is not readily transferrable between strains as plasmids are, the presence of K. pneumoniae in the normal human body and ease of international travel may allow the MDR strains of K. pneumoniae to spread faster than we can control.
Finally, the clonal distribution of K. pneumoniae tigecycline resistance in the Republic of Korea is dominated by several clonal types of K. pneumoniae, which have been linked to carbapenemases and other antibiotic resistances in the Republic of Korea.19 The six most common strains with tigecycline resistance, ST11, ST768, ST15, ST23, ST48, and ST307, comprise over 50% of the tigecycline-nonsusceptible cases in the Republic of Korea, and in particular, ST11. ST11 is a highly successful clonal type in East Asia2021 and also had been reported in association with tigecycline resistance in Spain in pets,11 although the resistance mechanism in that report had not been elucidated.
In conclusion, MDR K. pneumoniae, which are resistant to most known forms of antibiotics, occur nationwide in the Republic of Korea, and the development is led by several clonal types, which have been related to various antibiotic resistances in previous studies. Although there does not appear to be a transferrable method of nonsusceptibility at this point, over 16% of the tigecycline nonsusceptibility isolates arose by spontaneous mutation, seeming to suggest that this may be of concern in treating patients with the drug, especially since some of the clonal types found have been related with MDR in previous reports.
Figures and Tables
Table 1
Mutations Found in the ramR gene in Tigecycline Resistant K. pneumoniae Isolates

Table 2
Co-Resistance with Tigecycline
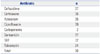
| Antibiotic | n |
|---|---|
| Ceftazidime | 37 |
| Ceftriaxone | 36 |
| Aztreonam | 35 |
| Ciprofloxacin | 39 |
| Carbapenems | 2 |
| Gentamicin | 17 |
| SXT | 32 |
| Tobramycin | 24 |
| Total | 56 |
Table 3
MLST Data of Tigecycline Nonsusceptible K. pneumoniae Isolates

ACKNOWLEDGEMENTS
This study was supported by a Grant from the Korean Health Technology R&D Project, Ministry of Health & Welfare, the Republic of Korea (A120843).
We are thankful to the Korean hospital network for providing the resistant bacterial isolates. We thank the team of the curators at the Institut Pasteur, MLST system (Paris, France) for importing novel alleles, profiles, and/or isolates at http://www.pasteur.fr/mlst. Tigecycline (Tigacyl™) 50 mg was kindly supplied by Pfizer, Korea.
References
1. Giamarellou H, Poulakou G. Multidrug-resistant Gram-negative infections: what are the treatment options? Drugs. 2009; 69:1879–1901.
2. Docobo-Pérez F, Nordmann P, Domínguez-Herrera J, López-Rojas R, Smani Y, Poirel L, et al. Efficacies of colistin and tigecycline in mice with experimental pneumonia due to NDM-1-producing strains of Klebsiella pneumoniae and Escherichia coli. Int J Antimicrob Agents. 2012; 39:251–254.

3. Lin YT, Wang FD, Chan YJ, Fu YC, Fung CP. Clinical and microbiological characteristics of tigecycline non-susceptible Klebsiella pneumoniae bacteremia in Taiwan. BMC Infect Dis. 2014; 14:1.
4. Sun Y, Cai Y, Liu X, Bai N, Liang B, Wang R. The emergence of clinical resistance to tigecycline. Int J Antimicrob Agents. 2013; 41:110–116.

5. Akiyama T, Presedo J, Khan AA. The tetA gene decreases tigecycline sensitivity of Salmonella enterica isolates. Int J Antimicrob Agents. 2013; 42:133–140.

6. Tuckman M, Petersen PJ, Projan SJ. Mutations in the interdomain loop region of the tetA(A) tetracycline resistance gene increase efflux of minocycline and glycylcyclines. Microb Drug Resist. 2000; 6:277–282.

7. Villa L, Feudi C, Fortini D, García-Fernández A, Carattoli A. Genomics of KPC-producing Klebsiella pneumoniae sequence type 512 clone highlights the role of ramR and ribosomal S10 protein mutations in conferring tigecycline resistance. Antimicrob Agents Chemother. 2014; 58:1707–1712.

8. Hentschke M, Wolters M, Sobottka I, Rohde H, Aepfelbacher M. ramR mutations in clinical isolates of Klebsiella pneumoniae with reduced susceptibility to tigecycline. Antimicrob Agents Chemother. 2010; 54:2720–2723.

9. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: 21st informational supplement (M100-S21). Wayne, PA: CLSI;2011.
10. Chiu SK, Wu TL, Chuang YC, Lin JC, Fung CP, Lu PL, et al. National surveillance study on carbapenem non-susceptible Klebsiella pneumoniae in Taiwan: the emergence and rapid dissemination of KPC-2 carbapenemase. PLoS One. 2013; 8:e69428.
11. Ovejero CM, Hidalgo L, Gutierrez B, Carrilero L, Santos-Lopez A, Thomas-Lopez D, et al. Human adapted Klebsiella pneumoniae ST11 and ST147 resistant to tigecycline from pet animal. In : Poster presentado en Med-Vet-Net Association International Scientific Conference 2013; 2013 Jun 24-25; Lyngby.
12. Rubio FG, Oliveira VD, Rangel RM, Nogueira MC, Almeida MT. Trends in bacterial resistance in a tertiary university hospital over one decade. Braz J Infect Dis. 2013; 17:480–482.

13. Spanu T, De Angelis G, Cipriani M, Pedruzzi B, D'Inzeo T, Cataldo MA, et al. In vivo emergence of tigecycline resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli. Antimicrob Agents Chemother. 2012; 56:4516–4518.

14. Landman D, Bratu S, Kochar S, Panwar M, Trehan M, Doymaz M, et al. Evolution of antimicrobial resistance among Pseudomonas aeruginosa, Acinetobacter baumannii and Klebsiella pneumoniae in Brooklyn, NY. J Antimicrob Chemother. 2007; 60:78–82.

15. Pakzad I, Zayyen Karin M, Taherikalani M, Boustanshenas M, Lari AR. Contribution of AcrAB efflux pump to ciprofloxacin resistance in Klebsiella pneumoniae isolated from burn patients. GMS Hyg Infect Control. 2013; 8:Doc15.
16. Bialek-Davenet S, Leflon-Guibout V, Tran Minh O, Marcon E, Moreau R, Nicolas-Chanoine MH. Complete deletion of the ramR gene in an in vitro-selected mutant of Klebsiella pneumoniae overexpressing the AcrAB efflux pump. Antimicrob Agents Chemother. 2013; 57:672–673.

17. Seecoomar GD, Marmol BC, Kwon DH. Promoter deletions of Klebsiella pneumoniae carbapenemase (KPC)-encoding genes (blaKPC-2) and efflux pump (AcrAB) on β-lactam susceptibility in KPC-producing Enterobacteriaceae. FEMS Microbiol Lett. 2013; 348:120–126.

18. Padilla E, Llobet E, Doménech-Sánchez A, Martínez-Martínez L, Bengoechea JA, Albertí S. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother. 2010; 54:177–183.

19. Shin SY, Bae IK, Kim J, Jeong SH, Yong D, Kim JM, et al. Resistance to carbapenems in sequence type 11 Klebsiella pneumoniae is related to DHA-1 and loss of OmpK35 and/or OmpK36. J Med Microbiol. 2012; 61(Pt 2):239–245.

20. Ko KS, Lee JY, Baek JY, Suh JY, Lee MY, Choi JY, et al. Predominance of an ST11 extended-spectrum beta-lactamase-producing Klebsiella pneumoniae clone causing bacteraemia and urinary tract infections in Korea. J Med Microbiol. 2010; 59(Pt 7):822–828.

21. Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011; 66:307–312.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download