Abstract
To characterize rotavirus G and P genotypes circulating among infants and young children hospitalized with severe diarrhea in a university hospital in Gyeonggi province, Korea, and to examine any association of the genotypes and nosocomial infections, we genotyped 103 isolates of rotavirus by multiplex RT-PCR. In July 2001-June 2002, we found that globally common strains constituted 64.2% (G2P[4] 28.3%, G3P[8] 28.3%, G4P[8] 5.7%, and G1P[8] 1.9%), and the uncommon strain, G4P[6], constituted 26.4%. During July 2002-June 2003, the percentage of common strains decreased to 44.0% (G3P[8] 18.0%, G2P[4] 16.8%, and G1P[8] 10.0%), but G4P[6] increased to 36.0%. G9P[8] was identified in 10.0% of cases, and thus can be considered an emerging strain in Korea. Eight-eight percent of G4P[6] was isolated from newborn babies. Among the 103 patients, there was an evidence of nosocomial rotavirus infection in 23 children (22.3%). Of these, 19 (82.6%) were newborns infected with G4P[6] strains of rotavirus. Most of the children who acquired rotavirus infection nosocomially showed symptoms of diarrhea, vomiting, fever, poor sucking, or dehydration, regardless of the genotype. This study revealed that G4P[6] has been the major genotype causing nosocomial rotavirus infection in our hospital.
Group A human rotavirus is a major etiologic agent of severe diarrhea in infants and young children in both developing and developed countries. Furthermore, it is the cause of death in approximately 600,000 children annually in developing countries (1, 2). Large scale studies in Korea have also shown that rotavirus was responsible for 24% to 46% of the hospitalized children with acute gastroenteritis (3, 4). Because of the large disease burden caused by rotavirus, prevention of rotavirus diarrhea is a major global health priority (1, 5). To design a vaccine strategy and to evaluate the success of candidate vaccines, it is essential to know the serotypes or genotypes of prevailing rotavirus strains in each country or region (6).
Group A rotaviruses possess a genome consisting of 11 segments of double-stranded RNA enclosed in a triple-layered capsid. The outer layer is composed of two proteins, VP4 and VP7, that elicit neutralizing antibody responses and that form the basis of the current dual classification system in G (VP7) and P (VP4) serotypes or genotypes. G serotypes have been identified by neutralization or enzyme-linked immunosorbent assay (ELISA), but genotyping of the G gene by nucleic acid amplification technique has been developed to facilitate rotavirus strain characterization (7). However serologic classification of P type has been more difficult because hyperimmune sera that distinguish individual P serotypes are not readily available. Instead, the nucleic acid amplification technique has been a reliable surrogate for the serotyping of P types (8).
Most of the studies conducted in Korea used ELISA to determine G serotypes and more than 90% of rotavirus isolates belonged to one of the four common G serotypes (G1-4) (3). More recently, both G and P genotyping studies have been reported in Korea (9-11). While conducting the rotavirus genotyping study in our institution, there have been many reports on outbreaks of rotavirus diarrhea in hospitals or in postpartum care facilities in Korea (12-14). But reports on association of rotavirus genotypes and nosocomial infections have been limited, so we tried to find any association of rotavirus genotypes and nosocomial infections.
Stool specimens from hospitalized pediatric patients with diarrhea were tested for rotavirus antigen by enzyme immunoassay (Vidas Rotavirus, bioMerieux sa, France). A total of 103 rotavirus-positive specimens (53 positive specimens in July 2001-June 2002 and 50 positives in July 2002-June 2003) were collected in Hanyang University Guri Hospital, in Gyeonggi province, Korea.
RNA was extracted with a commercially available mixture of phenol and guanidine thiocyanate (Tri Reagent LS, Molecular Research Center Inc., Cincinnati, OH, U.S.A.). Isolation of total RNA was performed according to the manufacture's instructions. Briefly, Tri Reagent LS was mixed with a 20% stool suspension in phosphate buffered saline (PBS) at a pH of 7.2 at a volume ratio of 3 to 1. After complete dissociation of nucleoprotein complexes, chloroform was added to the mixture followed by vigorous shaking and centrifugation at 12,000 g for 15 min at 4℃. The upper aqueous phase was transferred to a fresh tube and the RNA precipitated by mixing with isopropanol. The supernatant was removed and the RNA pellet was washed once with 75% ethanol. The RNA pellet was then briefly air-dried and dissolved in diethyl pyrocarbonate (DEPC) treated water.
RT-PCR methods for VP7 and VP4 genes are based on previously determined conditions used by Gentsch et al. (8) and Das et al. (15). Briefly, 1.2 µL of each of the primers, 9con1 and 9con2 for the VP7 gene (15), con3 and con2 for VP4 gene (8), and 5 µL RNA extract was added to the RT PreMix (Bioneer Inc., Daejeon, Korea), consisting of 4 µL 5 × RT buffer (250 mM Tris-HCl, 150 mM KCl, 40 mM MgCl2, pH 8.3), 2 µL 100 mM dithiothreitol, 0.5 µL deoxynucleoside triphosphate (dNTP) mix (dATP, dCTP, dGTP, and dTTP; each 10 mM), 20 units RNase inhibitor, and 200 units M-MLV RTase (Bioneer Inc.). The reaction mixture was then incubated at 42℃ for one hour. PCR PreMix (Bioneer Inc.) consisting of 1 unit Taq DNA polymerase, deoxynucleoside triphosphate (dNTP) mix (dATP, dCTP, dGTP, and dTTP; each 250 mM), 10 mM Tris-HCl (pH 9.0), 40 mM KCl, and 1.5 mM of MgCl2 was added to each tube, and then loaded into a preheated (94℃) Perkin Elmer thermal cycler (Gene Amp PCR System 9600, PerkinElmer Inc., Wellesley, MA, U.S.A.). PCR was performed as follows: 1 cycle at 94℃ for 5 min; 30 cycles at 94℃ for 0.5 min, 42℃ for 0.5 min, and 72℃ for 1 min; and 1 cycle at 72℃ for 7 min.
For G (VP7) genotyping, 1.2 µL of 10 µM 9con1 primer and each type-specific primer (9T-1, 9T-2, 9T-3P, 9T-4, 9T-9B) were used (15). For VP4 genotyping, 1.2 µL of 10 µM con3 primer and each type-specific primer (1-T1, 2-T1, 3-T1, 4-T1, 5T-1, ND2) were used (8). Conditions for the Nested PCR were identical to those used for the PCR described above. Five to ten µL of PCR products are analyzed on a 1.5% TBE agarose gel (Sigma-Aldrich, St. Louis, MO, U.S.A.) and viewed under ultraviolet illumination after ethidium bromide staining.
Individual patient records for children in whom rotavirus strains were genotyped were retrospectively reviewed to determine the presence of diarrhea (defined as three or more looser than normal stool passages within one day), fever (>38℃), nausea, vomiting, abdominal pain, and other symptoms such as poor oral intake.
Nosocomial rotavirus infection was defined as the presence of acute signs and symptoms of gastroenteritis (diarrhea, vomiting, fever, nausea, abdominal pain, or poor feeding), beginning 3 or more days following admission to the hospital for a non-diarrheal illness, and the presence of a rotavirus antigen in stool specimen (16).
From July 2001 to June 2003, a total of 1,031 fecal specimens were tested for rotavirus antigen at our hospital. During the two epidemic seasons of 2001-2002 and 2002-2003, 26.5% and 27.2% of stool specimens were positive for rotavirus. During the summer months of 2002 (June through August), the proportion of rotavirus-positive stool specimens was unusually high (27.7%). The 2002-2003 epidemic began on January 2003 and peaked from February through May 2003 (Table 1).
Among the 277 rotavirus antigen-positive specimens, we could genotype 103 strains (37.2%), and the rest of the specimens were too small in quantity to be genotyped. In the first year, globally common strains constituted 64.2%, while the uncommon strain, G4P[6] constituted 26.4%. During the second year, the proportion of common rotavirus strains declined to 44.0%. In 2002-2003, G4P[6] increased to 36.0% and globally emerging G9P[8] strain (10.0%) was detected (Table 2). One case of G4P[6] was identified in August 2001 and 11 additional cases were clustered during November 2001 through January 2002. In the 2002-2003 season, G4P[6] strains (n=18) were detected throughout the year. Analysis of the clinical pediatric data revealed that seven of 32 patients (21.8%) shed the G4P[6] rotavirus strain but did not have diarrhea. These seven children experienced relatively mild disease with loose stool, fever, or poor feeding. Eighty-eight percent (28/32) of G4P[6] strains were isolated from newborns. In contrast, 83.0% of the globally common strains (G2P[4] and G3P[8]) were isolated from children aged 6-35 months (Fig. 1).
Among the 103 patients whose charts were reviewed for the evidence of nosocomial infection, 23 (22.3%) were confirmed to have nosocomially-acquired rotavirus infection. Of the 23 children with confirmed nosocomial rotavirus infection, 19 (82.6%) were infected with G4P[6] strain and all were neonates (<30 days old). Other nosocomially acquired genotypes including one strain of G1P[8], one G2P[4], and two G3P[8] were found among children aged 2-42 months. Most infants who acquired rotavirus infection nosocomially showed symptoms of diarrhea, vomiting, fever, poor sucking, or dehydration regardless of genotype. Twelve children (52.2 %) were born by Cesarean section (Table 3).
Our two year study showed that the positive rate of rotavirus antigen among hospitalized children with gastroenteritis was 27% and this result is consistent with our previous ten year report (17) and a nationwide three year study in Korea (4). During this study period, the epidemic of rotavirus, which had begun in December 2001, continued through July 2002. This prolonged rotavirus epidemic that extended into the summer months (positive rate of 28%) was very unusual. Moreover, in 2003, a rotavirus peak appeared in the spring months (46% vs. 22% of winter season). Our previous study showed that the annual epidemic peaks of rotavirus diarrhea occurred in October, November, and December from 1989 through 1992. However, the peak time of rotavirus infection has gradually moved from December-January to February-March since 1993-94 (17). The underlying reasons for the change in the peak rotavirus season could not be explained by experimental evidence nor by errors in laboratory testing, but global warming could possibly influence the temperature in Korea and thus the rotavirus epidemic. Similar shifting of peak rotavirus activity from winter to spring has also been reported in Japan (18).
A recent report of global rotavirus strain collections revealed that four common G types (G1, G2, G3, and G4) in conjunction with P[8] or P[4] types represented over 88% of the rotavirus strains, but unusual or uncommon G and P types have been reported from various parts of the world (19). Recently, the G9 strain has been identified in a growing number of countries (20-24), and the neonatal type P[6] combined with G1, G3, G4, or G9 specificity, is emerging (25-28). The circulating strains of rotavirus differ by country and geographic region. In developed countries, the G1P[8] strain represents over 70% of rotavirus serotypes/genotypes, but only about 30% in South America and Asia, and 23% in Africa (19). P[6] represents almost one-third of all P types identified in Africa and 27% of the rotavirus infections were associated with strains bearing unusual combinations such as G8P[6] or G8P[4]. Previous serotyping studies of rotavirus conducted in Korea demonstrated that from 1987 until 1997, G1 was most prevalent strain and serotype G1-4 constituted more than 95% of serotyped strains (3). During the 1998-1999 epidemics, the G4 serotype became the most prevalent serotype, accounting for 58% of the serotyped strains. Recent reports from Korea revealed that G1, G2, G3, and G4 types constituted 96% (9), 87% (10), and 84% (11) of rotavirus strains.
In our study, the percentages of common G and P combined genotypes were different by year. The globally common G and P genotypes constituted 64.2% of infections in the 2001-2002 season, but decreased to 44.0% in the 2002-2003 season. The incidence of G2P[4] in the 2001-2002 season was quite high (28.3%) compared to the major global reports of 12% (19). The high incidence of uncommon genotypes was due mostly to the epidemic of G4P[6] strains (26.4% in 2001-2002 and 36.0% in 2002-2003), but in part due to the new emergence of G9P[8] strains since February 2003. The G9P[8] genotype, which has been a globally emergent strain, constituted 10% of specimens genotyped during 2002-2003. Overall, 54.4% of the strains genotyped for the two year period belonged to the globally common genotypes (Table 2).
The most intriguing result of our recent two year rotavirus genotyping study was the high incidence of the globally uncommon genotype, G4P[6] (31.1%), which is far higher than the results of Min et al. from Korea (10), and is actually the highest percentage of G4P[6] yet reported worldwide (19, 25). Although P[6] combined with G4 is considered rare globally and is reported to be associated with asymptomatic or symptomatic infections, P[6] strains combined with G1, G2, G3, or G4 have recently increased as a result of nosocomial rotavirus outbreaks in hospitals and newborn nurseries (25-29). There have been reports of rotavirus diarrhea epidemics in hospital nurseries and in postpartum care facilities in Korea since 2001 (12-14), and several hospitals even closed their newborn nurseries for a few months because it was so difficult to control the rotavirus transmission. From a hospital where a large epidemic of diarrhea in a newborn nursery occurred, we received 27 rotavirus antigen positive specimens (by latex agglutination) and genotyped them by RT-PCR. Except for one PCR-negative specimen, the remaining 26 were typed as G4P[6] (unpublished data). During the epidemic of G4P[6] rotavirus in newborn nurseries in Korea, many of the newborns suffered from severe diarrhea or even death in selected cases (14).
Our study revealed that among the 103 patients who shed rotavirus in their stool, there was evidence of nosocomial infection in 23 children (22.3%). Among the 23 nosocomially-infected children, 19 (82.6%) were infected with G4P[6] strains, all of whom were newborn babies. A hospital-based study over ten years in Germany reported nosocomial infection rate of rotavirus as being as high as 50% (29). Most studies to date have indicated that the neonatal rotavirus infection is predominantly asymptomatic, although severe diarrhea has been described in some instances (25). It remains unknown whether the attenuated infection in neonates is due to host factors or virulence factors of the neonatal rotavirus strains. In our study, most of the newborns positive for G4P[6] strains of rotavirus had symptoms (Table 3). This could be explained, in part, due to the selective testing of rotavirus for the symptomatic patients.
Although 92% of rotavirus strains isolated in our study belonged to the common G types, the newly emerging G9P[8] strains first appeared in Korea during February 2003. Therefore, a continued and nationwide genotype surveillance of rotavirus is needed to monitor changes in strain prevalence, to identify the emergence of new strains over time that could affect future vaccine strategies, and to identify any regional differences of genotype prevalence within Korea.
In summary, this study has extended our knowledge of rotavirus genotype diversity and highlights several important factors of rotavirus epidemiology in Korea. First, our study demonstrates that a globally rare strain, G4P[6], was the major epidemic strain in a university hospital in Korea. Secondly, the G4P[6] strain was the major cause of nosocomial rotavirus infection and was isolated mostly in newborn babies. Lastly, during the study period, the globally emerging strain G9P[8] was detected as a strain now emerging in Korea.
Figures and Tables
Fig. 1
Age distribution of commonly isolated rotavirus genotypes in Hanyang University Guri Hospital from July 2001 through June 2003.
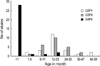
Table 1
Monthly percentages of rotavirus-positive specimens and peak months in Hanyang University Guri Hospital, July 2001 through June 2003

References
1. Wuethrich B, editor. Proceedings of the sixth international rotavirus symposium. 2005. 1st ed. Washington DC: The Albert B. Sabin Institute.
2. Parashar UD, Hummelman EG, Bresee JS, Miller MA, Glass RI. Global illness and deaths caused by rotavirus disease in children. Emerg Infect Dis. 2003. 9:565–572.

4. Kang JO, Kim MN, Kim J, Suh HS, Yoon Y, Jang S, Chang C, Choi S, Nyambat B, Kilgore PE. Epidemiologic trends of rotavirus infection in Republic of Korea, July 1999 through June 2002. Korean J Lab Med. 2003. 23:382–387.
5. Glass RI, Bresee JS, Parashar UD, Jiang B, Gentsch J. The future of rotavirus vaccines: a major setback leads to new opportunities. Lancet. 2004. 363:1547–1550.

6. Hoshino Y, Kapikian AZ. Rotavirus serotypes: Classification and importance in epidemiology, immunity, and vaccine development. J Health Popul Nutr. 2000. 18:5–14.
7. Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, Fang ZY. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990. 28:276–282.

8. Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, Das BK, Bhan MK. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992. 30:1365–1373.

9. Song MO, Kim KJ, Chung SI, Lim I, Kang SY, An CN, Kim W. Distribution of human group A rotavirus VP7 and VP4 types circulating in Seoul, Korea between 1998 and 2000. J Med Virol. 2003. 70:324–328.

10. Min BS, Noh YJ, Shin JH, Baek SY, Kim JO, Min KI, Ryu SR, Kim BG, Kim DK, Lee SH, Min HK, Ahn BY, Park SN. Surveillance study (2000 to 2001) of G- and P-type human rotaviruses circulating in South Korea. J Clin Microbiol. 2004. 42:4297–4299.

11. Kang JO, Kilgore P, Kim JS, Nyambat B, Kim J, Suh HS, Yoon Y, Jang S, Chang C, Choi S, Kim MN, Gentsch J, Bresee J, Glass R. Molecular epidemiological profile of rotavirus in South Korea, July 2002 through June 2003: Emergence of G4P[6] and G9P[8] strains. J Infect Dis. 2005. 192:Suppl 1. S57–S63.

12. Jang JM, Kim MJ, Cheong HW, Park DW, Sohn JW, Son CS, Lee SE. The epidemiologic characteristics and infection control measures for an outbreak of rotavirus infection in the neonatal unit. Infect Chemother. 2005. 37:311–318.
13. Korea CDC. Rotavirus detection in newborn babies in a day-care center in Goyang city, Gyeonggi province. CDMR. 2003. 14:284.
14. Yang KM, Park SH, Kim IS, Lee JH, Lee HY, Kwon TJ, Lee WT, Kang SM, Yang BK, Goh UY, Ji YM. Report of newborn deaths at post-delivery care facilities in 2001 and 2002. Korean J Leg Med. 2002. 26:33–46.
15. Das BK, Gentsch JR, Cicirello HG, Woods PA, Gupta A, Ramachandran M, Kumar R, Bhan MK, Glass RI. Characterization of rotavirus strains from newborns in New Delhi, India. J Clin Microbiol. 1994. 32:1820–1822.

16. Farr BM. Mayhall CG, editor. Nosocomial gastrointestinal tract infections. Hospital Epidemiology and Infection Control. 1999. 2nd ed. Philadelphia, USA: Lippincott Willams & Wilkins;247–274.
17. Kang JO, Kim SE, Kim TY, Park IK, Choi TY. Trends in rotavirus gastroenteritis in Korea from 1989 through 1998 and comparison of Slidex Rota-kit 2 and VIDAS Rotavirus. Korean J Clin Microbiol. 1999. 2:152–155.
18. Suzuki H, Sakai T, Tanabe N, Okabe N. Peak rotavirus activity shifted from winter to early spring in Japan. Pediatr Infect Dis J. 2005. 24:257–260.

19. Santos N, Hoshino Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol. 2005. 15:29–56.

20. Unicomb LE, Podder G, Gentsch JR, Woods PA, Hasan KZ, Faruque AS, Albert MJ, Glass RI. Evidence of high-frequency genomic reassortment of group A rotavirus strains in Bangladesh: Emergence of type G9 in 1995. J Clin Microbiol. 1999. 37:1885–1891.
21. Steele AD, Ivanoff B. Rotavirus strains circulating in Africa during 1996-1999: emergence of G9 strains and P[6] strains. Vaccine. 2003. 21:361–367.

22. Clark HF, Lawley DA, Schaffer A, Patacsil JM, Marcello AE, Glass RI, Jain V, Gentsch J. Assessment of the epidemic potential of a new strain of rotavirus associated with the novel G9 serotype which caused an outbreak in the United States for the first time in the 1995-1996 season. J Clin Microbiol. 2004. 42:1434–1438.

23. Santos N, Volotao EM, Soares CC, Albuquerque MC, da Silva FM, de Carvalho TR, Pereira CF, Chizhikov V, Hoshino Y. Rotavirus strain bearing genotype G9 or P[9] recovered form Brazilian children with diarrhea from 1997 to 1999. J Clin Microbiol. 2001. 39:1157–1160.
24. Griffin DD, Kirkwood CD, Parashar UD, Woods PA, Bresee JS, Glass RI, Gentsch JR. Surveillance of rotavirus strains in the United States: Identification of unusual strains. J Clin Microbiol. 2000. 38:2784–2787.
25. Gentsch JR, Woods PA, Ramachandran M, Das BK, Leite JP, Alfieri A, Kumar R, Bhan MK, Glass RI. Review of G and P typing results from a global collection of rotavirus strains: implications for vaccine development. J Infect Dis. 1996. 174:Suppl 1. S30–S36.

26. Widdowson MA, van Doornum GJ, van der Poel WH, de Boer AS, Mahdi U, Koopmans M. Emerging group-A rotavirus and a nosocomial outbreak of diarrhoea. Lancet. 2000. 356:1161–1162.

27. Lee CN, Lin CC, Kao CL, Zao CL, Shih MC, Chen HN. Genetic characterization of the rotaviruses associated with a nursery outbreak. J Med Virol. 2001. 63:311–320.

28. Linhares AC, Mascarenhas JD, Gusmao RH, Gabbay YB, Fialho AM, Leite JP. Neonatal rotavirus infection in Belem, Nothern Brazil: Nosocomial transmission of a P[6]G2 strains. J Med Virol. 2002. 67:418–426.
29. Berner R, Schumacher RF, Hameister S, Forster J. Occurrence and impact of community-acquired and nosocomial rotavirus infections--a hospital-based study over 10 y. Acta Paediatr Suppl. 1999. 426:48–52.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download