Abstract
Background
Multidrug-resistant Enterobacteriaceae is a worldwide problem. Although various resistance mechanisms have been recognized with increasing frequency, only a few cases of triple resistance of extended-spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae have been reported. This study was designed to evaluate the coexistence of qnr (qnrA, qnrB, and qnrS) and 16S rRNA methylase (armA, rmtA, rmtB, and rmtC) in ESBL-producing K. pneumoniae.
Methods
We tested 44 isolates of ESBL-producing K. pneumoniae at Chungnam National University Hospital from March to September 2006. Antimicrobial susceptibilities were tested by broth microdilution method, and transconjugation test was performed using E. coli J53 with azide resistance. Search for qnr (qnrA, qnrB, and qnrS) and 16S rRNA methylase (armA, rmtA, rmtB, and rmtC) genes was conducted by PCR amplification, and the genotypes were determined by direct nucleotide sequence analysis of the amplified products. Epidemiologic study was performed by Enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR).
REFERENCES
1.Abbott S. Klebsiella, Enterobacter, Citrobacter and Serratia. Murray PR, Baron EJ, editors. Manual of Clinical Microbiology. 7th ed.Washington: ASM press;1999. p. 475–82.
2.Podschum R., Ullman U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998. 11:589–603.
3.Lee YH., Cho B., Bae IK., Chang CL., Jeong SH. Klebsiella pneumoniae strains carrying the chromosomal SHV-11 β-lactamase more frequently than those carrying the chromosomal SHV-1 β-lactamase gene. J Antimicrob Chemother. 2006. 57:1259–61.
4.Quitilaiani R Jr., Sahm DF, et al. Mechanisms of resistance to antimicrobial agents. Murray PR, Baron EJ, editors. Manual of Clinical Microbiology. 7th ed.Washington: ASM press;1999. p. 1505–25.
5.Shin KS., Son BR. Comparison of Vitek ESBL test and other methods for detecting extended-spectrum β-lactamase producing Escherichia coli and Klebsiella species. Korean J Clin Pathol. 2002. 22:21–6. (신경섭및손보라. Escherichia coli와 Klebsiella species에서 Extended-spectrum -lactamase 검출을위한 Vitel ESBL test와그외방법의비교. 대한임상병리학회지 2002;22: 21-6.).
6.Hong SG., Kang MS., Choi JR., Lee KW., Chong YS., Kwon OH. Molecular characteristics of extended-spectrum β-lactamases in clinical isolates of enterobacteriaceae. Korean J Clin Pathol. 2001. 21:495–504. (홍성근, 강명서, 최종락, 이경원, 정윤섭, 권오헌. 임상검체에서 분리된 Enterobacteriaceae 균종의 Extended-spectrum -lactamase 유형및분자역학적특성. 대한임상병리학회지 2001;21: 495-504.).
7.Lee BY., Jeong SH., Jeong TS., Nam HJ., Ji JH., Hong YR. Detection of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella spp. with the Vitek GNS 121 card. Korean J Clin Pathol. 2001. 21:350–4. (이보영, 정석훈, 정태식, 남희준, 지종현, 홍유라. Vitek GNS 121 Card를이용한 Extended-spectrum -Lactamase 생성 Escherichia coli와 Klebsiella spp. 검출. 대한임상병리학회지 2001;21: 350-4.).
8.Lautenbach E., Strom BL., Nachamkin I., Bilker WB., Marr AM., Larosa LA, et al. Longitudinal trends in fluoroquinolone resistance among Enterobacteriacae isolates from inpatients and outpatients, 1989-2000: differences in the emergence and epidemiology of resistance across organisms. Clin Infect Dis. 2004. 38:655–62.
9.Jacoby GA., Chow N., Waites KB. Prevalence of plasmid-mediated quinolone resistance. Antimicrob Agents Chemother. 2003. 47:559–62.

10.Wang M., Sahm DF., Jacoby GA., Hooper DC. Emerging plasmid-mediated quinolone resistance associated with qnr gene in Klebsiella pneumoniae clinical isolates in the United States. Antimicrob Agents Chemother. 2004. 48:1295–9.
11.Itokazu GS., Quinn JP., Bell-Dixon C., Kahan FM., Weinstein RA. Antimicrobial resistance rates among aerobic gram-negative bacilli recovered from patients in intensive care units: evaluation of a national postmarketing surveillance program. Clin Infect Dis. 1996. 23:779–84.

12.Lautenbach E., Strom BL., Bilker WB., Patel JB., Edelstein PH., Fishman NO. Epidemiological investigation of fluoroquinolone resistance in infections due to extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Clin Infect Dis. 2001. 33:1288–94.
13.Paterson DL., Mulazimoglu L., Casellas JM., Ko WC., Goossens H., Von Gottberg A, et al. Epidemiology of ciprofloxacin resistance and its relationship to extended-spectrum β-lactamase production in Klebsiella pneumoniae isolates causing bacteremia. Clin Infect Dis. 2000. 30:473–8.
14.Kotra LP., Haddad J., Mobashery S. Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicorb Agents Chemother. 2000. 44:3249–56.

15.Galimand M., Courvalin P., Lambert T. Plasmid-mediated high level resistance to aminoglycosides in Enterobacteriaceae due to 16s rRNA methylation. Antimicrob Agents Chemother. 2003. 47:2565–71.
16.Galimand M., Sabtcheva S., Courvalin P., Lambert T. Worldwide disseminated armA aminoglycoside resistance methlyase gene is borne by composite transposon Tn1548. Antimicrob Agents Chemother. 2005. 49:2949–53.
17.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; sixteenth informational supplement. M100-S16. Wayne, Pensylvania: CLSI;2006.
18.Jacoby GA., Han P. Detection of extended-spectrum beta-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol. 1996. 34:908–11.
19.Versalovic J., Koeuth T., Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1999. 19:6823–31.
20.Livermore DM. β-Lactamases in laboratory and clinical? resistance. Clin Microbioal Rev. 1995. 8:557–84.
21.Hong SG., Kim SJ., Jeong SH., Chang CH., Cho SR., Ahn JY, et al. Prevalence and diversity of extened-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates in Korea. Korean J Clin Microbiol. 2003. 6:149–55. (홍성근, 김선주, 정석훈, 장철훈, 조성란, 안지영 등. 국내에서 분리된 Extended-spectrum -lactamase 생성 Escherichia coli and Klebsiella pneumoniae의 빈도 및 유형. 대한임상미생물학회지 2003;6: 149-55.).
22.Wang M., Tran JH., Jacoby GA., Zhang Y., Wang F., Hooper DC. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob Agents Chemother. 2003. 47:2242–8.
23.Bogaerts P., Galimand M., Baurang C., Deplano A., Vanhoof R., De Mendonca R, et al. Emergence of ArmA and RmtB aminoglycoside resistance 16S rRNA methylases in Belgium. J Antimicrob Chemother. 2007. 59:459–64.
24.Martinez-Martinez L., Pascual A., Garcia I., Tran J., Jacoby GA. Interaction of plasmid and host quinolone resistance. J Antimicrob Chemother. 2003. 51:1037–9.

25.Yan JJ., Wu JJ., Ko WC., Tsai SH., Chuang CL., Wu HM, et al. Plasmid-mediated 16S rRNA methylase conferring high-level aminoglycoside resistance in Escherichia coli and Klebsiella pneumoniae isolates from two Taiwanese hospitals. J Antimicrobial Chemother. 2004. 54:1007–12.
26.Lee H., Yong D., Yum JH., Roh KH., Lee K., Yamane K, et al. Dissemination of 16S rRNA methylase-mediated highly amikacin-resistant isolates of Klebsiella pneumoniae and Acinetobacter baumannii in Korea. Diagn Microbiol Infect Dis. 2006. 56:305–12.
27.Lee K., Lee M., Shin JH., Lee MH., Kang SH., Park AJ, et al. Prevalence of plasmid-mediated AmpC β-lactamases in Escherichia coli and Klebsiella pneumoniae in Korea. Microb Drug Resist. 2006. 12:44–9.
28.Kang JH., Bae IK., Kwon SB., Jeong SH., Lee JW., Lee WG, et al. Prevalence of Ambler Class A extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates in Korea. Korean J Clin Microbiol. 2005. 8:17–25. (강지혜, 배일권, 권수봉, 정석훈, 이종욱, 이위교등. Ambler Class A Extended-Spectrum -Lactamase 생성 Escherichia coli와 Klebsiella pneumoniae의국내분리현황. 대한임상미생물학회지 2005;8: 17-25.).
29.Cattoir V., Weill FX., Poirel L., Fabre L., Soussy CJ., Nordmann P. Prevalence of qnr genes in Salmonella in France. J Antimicrob Chemother. 2007. 59:751–4.
30.Park YJ., Lee S., Yu JK., Woo GJ., Lee K., Arakawa Y. Co-production of 16S rRNA methylases and extended-spectrum beta-lactamases in AmpC-producing Enterobacter cloacae, Citrobacter freundii and Serratia marcescens in Korea. J Antimicrob Chemother. 2006. 58:907–8.
Fig. 1.
Enterobacterial repetitive intergenic consensus-PCR patterns of genomic DNA from clinical isolates of K. pneumoniae harboring ESBLs. Lane M is 1kb DNA size marker. Among the five different banding patterns (A, B, C, D, E), A and B patterns are dominant. Thirty nine of 44 extended-spectrum β-lactamase -producing clinical isolates show A or B patterns.
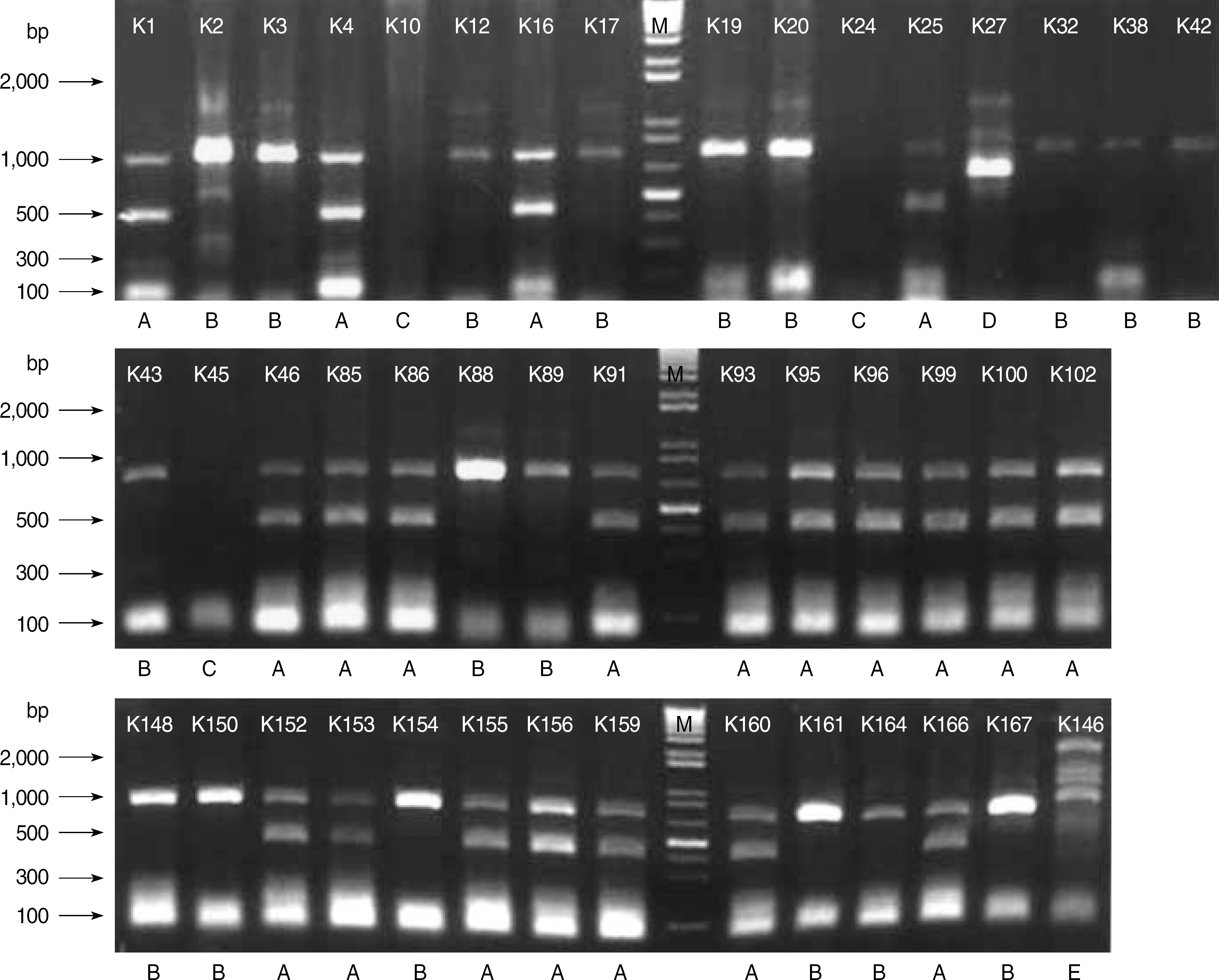
Table 1.
Oligonucleotides used as primers for amplification and sequencing in this study
| Enzymes | Primer pairs | Target | Sequence (5′-3′) | Amplicon size (bp) | Reference |
|---|---|---|---|---|---|
| β-lactamase | TEM F∗ | blaTEM and variants | ATGAGTATTCAACATTTCCGT | 861 | 28 |
| TEM R† | TTACCAATGCTTAATCAGTGA | ||||
| SHV F | blaSHV and variants | CCGGGTTATTCTTATTTGTCGCT | 831 | 28 | |
| SHV R | TAGCGTTGCCAGTGCTCG | ||||
| CTX-M1F | blaCTX-M-1 cluster | AGTTCACGCTGATGGCGACG | 676 | 23 | |
| CTX-M1R | AACCCAGGAAGCAGGCAGTCC | ||||
| CTX-M9F | blaCTX-M-9 cluster | GATTGACCGTATTGGGAGTTT | 947 | 28 | |
| CTX-M9R | CGGCTGGGTAAAATAGGTCA | ||||
| Qnr | qnrA F | qnrA | GGGTATGGATATTATTGATAAAG | 660 | 29 |
| qnrA R | CTAATCCGGCAGCACTATTA | ||||
| qnrB F | qnrB1-qnrB6 variants | GGMATHGAAATTCGCCACTG | 264 | 29 | |
| qnrB R | TTTGCYGYYCGCCAGTCGAA | ||||
| qnrS F | qnrS | AGTGATCTCACCTTCACCGC | 550 | 29 | |
| qnrS R | CAGGCTGCAATTTTGATACC | ||||
| 16S rRNA methylase | armA F | armA | AGGTTGTTTCCATTTCTGAG | 776 | 23 |
| armA R | TCTCTTCCATTCCCTTCTCC | ||||
| rmtA F | rmtA | CTAGCGTCCATCCTTTCCTC | 635 | 23 | |
| rmtA R | TTTGCTTCCATGCCCTTGCC | ||||
| rmtB F | rmtB | CCCAAACAGACCGTAGAGGC | 769 | 23 | |
| rmtB R | CTCAAACTCGGCGGGCAAGC | ||||
| rmtC F | rmtC | CGAAGAAGTAACAGCCAAAG | 700 | 30 |
Table 2.
Characteristics of ESBL-producing K. pneumoniae harboring qnrB, qnrS, and armA genes
Table 3.
Activities of antimicrobial agents against ESBL producing K. pneumoniae harboring qnrB, qnrS, and armA genes
Table 4.
Distributions of qnrB, qnrS and armA determinants according to ESBL among clinical isolates of K. pneumoniae
Table 5.
Resistance profile of donor strains and transconjugants




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download