Dear Editor,
9p duplication syndrome is characterized by craniofacial dysmorphism, digital abnormalities, short stature, short neck, developmental delay, and mental retardation [1]. It can also accompany rare phenotypes including cardiac defect [23], psychotic behavior [4], autism spectrum disorder (ASD) [5], and hearing loss [6]. Phenotypes of 9p duplication correlate with the size and position of the involved region [6]. Most 9p duplications originate from parental balanced translocation, usually between chromosome 9 and other autosomes [14]. Therefore, accompanying partial monosomy of chromosome 9 or abnormalities of other chromosomes may complicate the understanding on the genotype-phenotype correlation of 9p duplication [1]. Reports on isolated 9p duplication, excluding cases with partial monosomy of chromosome 9 or other chromosomes, are relatively uncommon [1]. Here, we report an isolated 9p duplication case confirmed by chromosomal microarray (CMA) analysis.
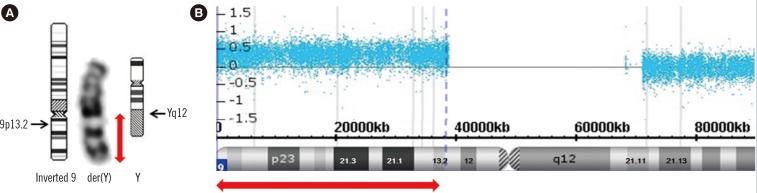
The patient was a 38-yr-old Korean man with severe mental retardation and no other underlying disease previously diagnosed. On the day of admission, he had a sudden cardiac arrest due to ventricular fibrillation at his work facility. Further evaluation after resuscitation and admission revealed severe aortic stenosis from bicuspid aortic valve. Tracheomalacia was noted by fiber-optic bronchoscopy (FOB). Brain computed tomography found decreased brain volume and mild hydrocephalus. On the basis of his dysmorphic features (coarse face and short neck) and cardiac anomaly, the attending clinician suspected DiGeorge syndrome, but FISH targeting the TUPLE1 gene (Abbott Molecular, Downers Grove, IL, USA) showed negative results. In G-banded karyotyping, material of unknown origin was inserted into the heterochromatin portion of the Y chromosome (Fig. 1A). CMA analysis with a CytoScan 750K array (Affymetrix, Santa Clara, CA, USA) revealed a 38.5 mega base (Mb) duplication on 9p24.3p13.2 (Fig. 1B). The final cytogenetic result for the patient was reported as 46,X,der(Y)t(Y;9)(q12;p13.2).arr[hg19] 9p24.3p13.2(208,454-38,689,749)x3 according to the International System for Human Cytogenetic Nomenclature 2013.
The duplicated region in our patient includes a critical region of 9p duplication syndrome (9p22.3 to 9p22.2) [5] and contains 255 genes and 147 Online Mendelian Inheritance in Man (OMIM) genes. Among them, DOCK8, KANK1, VLDLR, MLLT3, and PIGO are known to be associated with mental retardation or developmental delay, usually involving deletion, translocation breakpoint, or point mutation of these genes (http://www.ncbi.nlm.nih.gov/omim).
Congenital tracheomalacia can be associated with chromosomal abnormality [7], but no 9p duplication case has been reported in the literature. From the DatabasE of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER, https://decipher.sanger.ac.uk/), we retrieved over 180 cases having duplicated regions overlapping with our case. Among them, only one patient (ID: 282249) with a 749 kb size duplication at 9p13.3 had tracheomalacia along with developmental delay and hypothyroidism. The frequency could be underestimated since mild tracheomalacia without associated tracheoesophageal anomaly cannot be identified by routine clinical evaluation. We performed FOB in our patient because of desaturation despite the intubation condition, and without this event, we might have missed his tracheomalacia.
Abu-Amero et al. [5] suggested that 9p23-24.3 can be a potential ASD locus. However, there are many cases involving 9p23-24.3 but without ASD, and instead, all including our case had developmental delay and/or mental retardation [1456]. The explanation for such a variation may include a combinatorial effect of various factors including other genetic variants, epigenetic regulation, and environmental factors [8].
Although cardiac defect is uncommon in 9p duplication, three such cases have been reported including dup(9)(p22 p24), supernumerary der(9)(pter→q13::q13→q12:), and der (9)t(9;21)(q13;q21) [239]. From the DECIPHER database, we found two more cases with 9p duplication and cardiac defects, including a case with a 346-kb duplication at 9p24.1 and a case with a 614-kb duplication at 9p13.3 (IDs: 256847 and 276350, respectively). Nonetheless, the pathogenicity and consensus on the critical chromosomal region are yet uncertain, reflecting the heterogeneous nature of causative loci for cardiac defects in the human genome [10].
In Korea, six cases of 9p duplication syndrome have been reported until now [9]. All previous cases were diagnosed by using G-banded karyotyping, FISH, or multiplex ligation-dependent probe amplification. Here we report the seventh Korean case of 9p duplication, for which CMA analysis was firstly used for the diagnosis of this syndrome in Korea. In cases with unidentifiable derivative chromosomes by conventional karyotyping, CMA can be indispensable for the identification of the abnormalities, the breakpoint, and altered genes.
Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2012R1A1A2043879).
References
1. Guilherme RS, Meloni VA, Perez AB, Pilla AL, de Ramos MA, Dantas AG, et al. Duplication 9p and their implication to phenotype. BMC Med Genet. 2014; 15:142. PMID: 25526829.

2. Teraoka M, Narahara K, Yokoyama Y, Ninomiya S, Mizuta S, Une T, et al. Maternal origin of a unique extra chromosome, der(9)(pter→q13:: q13→q12:) in a girl with typical trisomy 9p syndrome. Am J Med Genet. 2001; 102:25–28. PMID: 11471168.
3. Haddad BR, Lin AE, Wyandt H, Milunsky A. Molecular cytogenetic characterisation of the first familial case of partial 9p duplication (p22p24). J Med Genet. 1996; 33:1045–1047. PMID: 9004142.

4. Martínez-Jacobo L, Ortíz-López R, Rizo-Méndez A, García-Molina V, Santuario-Facio SK, Rivas F, et al. Clinical and molecular delineation of duplication 9p24.3q21.11 in a patient with psychotic behavior. Gene. 2015; 560:124–127. PMID: 25667990.

5. Abu-Amero KK, Hellani AM, Salih MA, Seidahmed MZ, Elmalik TS, Zidan G, et al. A de novo marker chromosome derived from 9p in a patient with 9p partial duplication syndrome and autism features: genotype-phenotype correlation. BMC Med Genet. 2010; 11:135. PMID: 20858261.

6. Zhou YC, Zhang C, Zhai JS, Li TF, Wu QY, Li WW, et al. A patient with unusual features and a 69.5 Mb duplication from a de novo extra der (9): a case report. Mol Med Rep. 2015; 12:155–158. PMID: 25760145.

7. Wilson MG, Towner JW, Forsman I, Siris E. Syndromes associated with deletion of the long arm of chromosome 18[del(18q)]. Am J Med Genet. 1979; 3:155–174. PMID: 474629.

8. Cooper DN, Krawczak M, Polychronakos C, Tyler-Smith C, Kehrer-Sawatzki H. Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum Genet. 2013; 132:1077–1130. PMID: 23820649.

9. Woo KS, Kim KE, Kwon EY, Kim JP, Han JY. A case of partial trisomy 9pter→q13 due to paternal balanced translocation t (9;21) (q13;q21). Korean J Lab Med. 2008; 28:155–159. PMID: 18458513.

10. Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature. 2006; 444:444–454. PMID: 17122850.

Fig. 1
G-banded karyotype and chromosomal microarray results of the patient. (A) G-banding revealed inserted material on the Y chromosome (red arrow). The karyotype was confirmed as 46,X,der(Y)(Ypter→Yq12::9p13.2→9pter) after microarray analysis. (B) The microarray results showed a duplication in the 9p24.3p13.2 (chr9:208,454-38,689,749) region (red arrow). Log2 value (blue dots) of 0.5 in the region represents a 3:2 copy number ratio of the test to the reference DNA.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download