1. Perez LE. Outcomes from unrelated donor hematopoietic stem cell transplantation. Cancer Control. 2011; 18:216–221. PMID:
21976240.

2. Shaw BE, Arguello R, Garcia-Sepulveda CA, Madrigal JA. The impact of HLA genotyping on survival following unrelated donor haematopoietic stem cell transplantation. Br J Haematol. 2010; 150:251–258. PMID:
20560963.

3. Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002; 295:2097–2100. PMID:
11896281.

4. Malmberg KJ, Schaffer M, Ringdén O, Remberger M, Ljunggren HG. KIR-ligand mismatch in allogeneic hematopoietic stem cell transplantation. MolImmunol. 2005; 42:531–534.

5. Ruggeri L, Mancusi A, Burchielli E, Aversa F, Martelli MF, Velardi A. Natural killer cell alloreactivity in allogeneic hematopoietic transplantation. Curr Opin Oncol. 2007; 19:142–147. PMID:
17272987.

6. Petersdorf EW, Gooley T, Malkki M, Horowitz M. International Histocompatibility Working Group in Hematopoietic Cell Transplantation. Clinical significance of donor-recipient HLA matching on survival after myeloablative hematopoietic cell transplantation from unrelated donors. Tissue Antigens. 2007; 69(S1):25–30. PMID:
17445158.

7. Morishima Y, Kawase T, Malkki M, Petersdorf EW. International Histocompatibility Working Group in Hematopoietic Cell Transplantation Component. Effect of HLA-A2 allele disparity on clinical outcome in hematopoietic cell transplantation from unrelated donors. Tissue Antigens. 2007; 69(S1):31–35. PMID:
17445159.

8. Lee KW, Oh DH, Lee C, Yang SY. Allelic and haplotypic diversity of HLA-A, -B, -C, -DRB1, and -DQB1 genes in the Korean population. Tissue Antigens. 2005; 65:437–447. PMID:
15853898.

9. Yang JH, Sohn YH, Ko SY, Choi SE, Kim MH, Oh HB. Anthropological analysis of Koreans using HLA class II diversity among East Asians. Tissue Antigens. 2010; 76:282–288. PMID:
20522202.

10. Farag SS, Fehniger TA, Ruggeri L, Velardi A, Caligiuri MA. Natural killer cell receptors: new biology and insights into the graft-versus-leukemia effect. Blood. 2002; 100:1935–1947. PMID:
12200350.

11. Giebel S, Locatelli F, Lamparelli T, Velardi A, Davies S, Frumento G, et al. Survival advantage with KIR ligand incompatibility in hematopoietic stem cell transplantation from unrelated donors. Blood. 2003; 102:814–819. PMID:
12689936.

12. Glucksberg H, Storb R, Fefer A, Buckner CD, Neiman PE, Clift RA, et al. Clinical manifestations of graft-versus-host disease in human recipients of marrow from HL-A-matched sibling donors. Transplantation. 1974; 18:295–304. PMID:
4153799.

13. Gratwohl A. The EBMT risk score. Bone Marrow Transplant. 2012; 47:749–756. PMID:
21643021.

14. Morishima Y, Sasazuki T, Inoko H, Juji T, Akaza T, Yamamoto K, et al. The clinical significance of human leukocyte antigen (HLA) allele compatibility in patients receiving a marrow transplant from serologically HLA-A, HLA-B, and HLA-DR matched unrelated donors. Blood. 2002; 99:4200–4206. PMID:
12010826.

15. Kawase T, Morishima Y, Matsuo K, Kashiwase K, Inoko H, Saji H, et al. High-risk HLA allele mismatch combinations responsible for severe acute graft-versus-host disease and implication for its molecular mechanism. Blood. 2007; 110:2235–2241. PMID:
17554059.

16. Lee SJ, Klein J, Haagenson M, Baxter-Lowe LA, Confer DL, Eapen M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007; 110:4576–4583. PMID:
17785583.

17. Hsu KC, Gooley T, Malkki M, Pinto-Agnello C, Dupont B, Bignon JD, et al. KIR ligands and prediction of relapse after unrelated donor hematopoietic cell transplantation for hematologic malignancy. Biol Blood Marrow Transplant. 2006; 12:828–836. PMID:
16864053.

18. Giebel S, Locatelli F, Wojnar J, Velardi A, Mina T, Giorgiani G, et al. Homozygosity for human leucocyte antigen-C ligands of KIR2DL1 is associated with increased risk of relapse after human leucocyte antigen-C-matched unrelated donor haematopoietic stem cell transplantation. Br J Haematol. 2005; 131:483–486. PMID:
16281939.

19. Fischer JC, Ottinger H, Ferencik S, Sribar M, Punzel M, Beelen DW, et al. Relevance of C1 and C2 epitopes for hemopoietic stem cell transplantation: role for sequential acquisition of HLA-C-specific inhibitory killer Ig-like receptor. J Immunol. 2007; 178:3918–3923. PMID:
17339492.

20. Fischer JC, Kobbe G, Enczmann J, Haas R, Uhrberg M. The impact of HLA-C matching depends on the C1/C2 KIR ligand status in unrelated hematopoietic stem cell transplantation. Immunogenetics. 2012; 64:879–885. PMID:
22923051.

21. Ludajic K, Balavarca Y, Bickeböller H, Rosenmayr A, Fae I, Fischer GF, et al. KIR genes and KIR ligands affect occurrence of acute GVHD after unrelated, 12/12 HLA matched, hematopoietic stem cell transplantation. Bone Marrow Transplant. 2009; 44:97–103. PMID:
19169284.

22. Gagne K, Busson M, Bignon JD, Balère-Appert ML, Loiseau P, Dormoy A, et al. Donor KIR3DL1/3DS1 gene and recipient Bw4 KIR ligand as prognostic markers for outcome in unrelated hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2009; 15:1366–1375. PMID:
19822295.

23. Miller JS, Cooley S, Parham P, Farag SS, Verneris MR, McQueen KL, et al. Missing KIR ligands are associated with less relapse and increased graft-versus-host disease (GVHD) following unrelated donor allogeneic HCT. Blood. 2007; 109:5058–5061. PMID:
17317850.

24. Farag SS, Bacigalupo A, Eapen M, Hurley C, Dupont B, Caligiuri MA, et al. The effect of KIR ligand incompatibility on the outcome of unrelated donor transplantation: a report from the center for international blood and marrow transplant research, the European blood and marrow transplant registry, and the Dutch registry. Biol Blood Marrow Transplant. 2006; 12:876–884. PMID:
16864058.

25. Willemze R, Ruggeri A, Purtill D, Rodrigues CA, Gluckman E, Rocha V, et al. Is there an impact of killer cell immunoglobulin-like receptors and KIR-ligand incompatibilities on outcomes after unrelated cord blood stem cell transplantation? Best Pract Res Clin Haematol. 2010; 23:283–290. PMID:
20837340.

26. Hollenbach JA, Augusto DG, Alaez C, Bubnova L, Fae I, Fischer G, et al. 16(th) IHIW: population global distribution of killer immunoglobulin-like receptor (KIR) and ligands. Int J Immunogenet. 2013; 40:39–45. PMID:
23280119.
27. Shilling HG, Young N, Guethlein LA, Cheng NW, Gardiner CM, Tyan D, et al. Genetic control of human NK cell repertoire. J Immunol. 2002; 169:239–247. PMID:
12077250.

28. Nguyen S, Dhedin N, Vernant JP, Kuentz M, Al Jijakli A, Rouas-Freiss N, et al. NK-cell reconstitution after haploidentical hematopoietic stem-cell transplantations: immaturity of NK cells and inhibitory effect of NKG2A override GvL effect. Blood. 2005; 105:4135–4142. PMID:
15687235.

29. Stern M, de Angelis C, Urbani E, Mancusi A, Aversa F, Velardi A, et al. Natural killer-cell KIR repertoire reconstitution after haploidentical SCT. Bone Marrow Transplant. 2010; 45:1607–1610. PMID:
20173785.

30. Parham P. Immunology.NK cells lose their inhibition. Science. 2004; 305:786–787. PMID:
15297654.
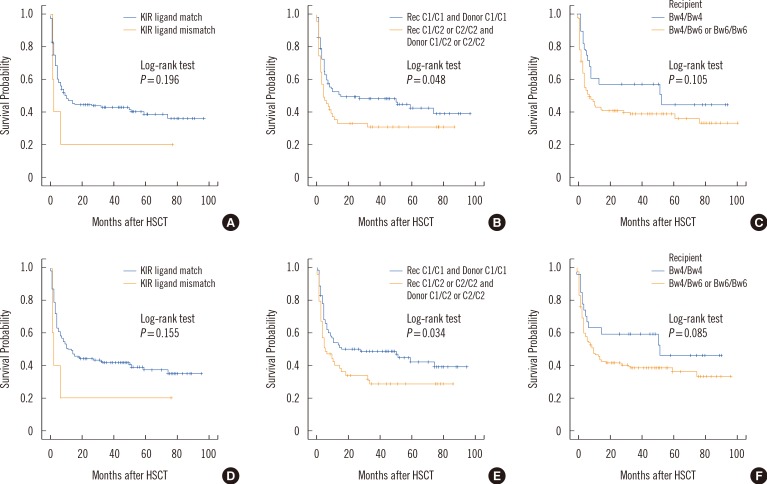
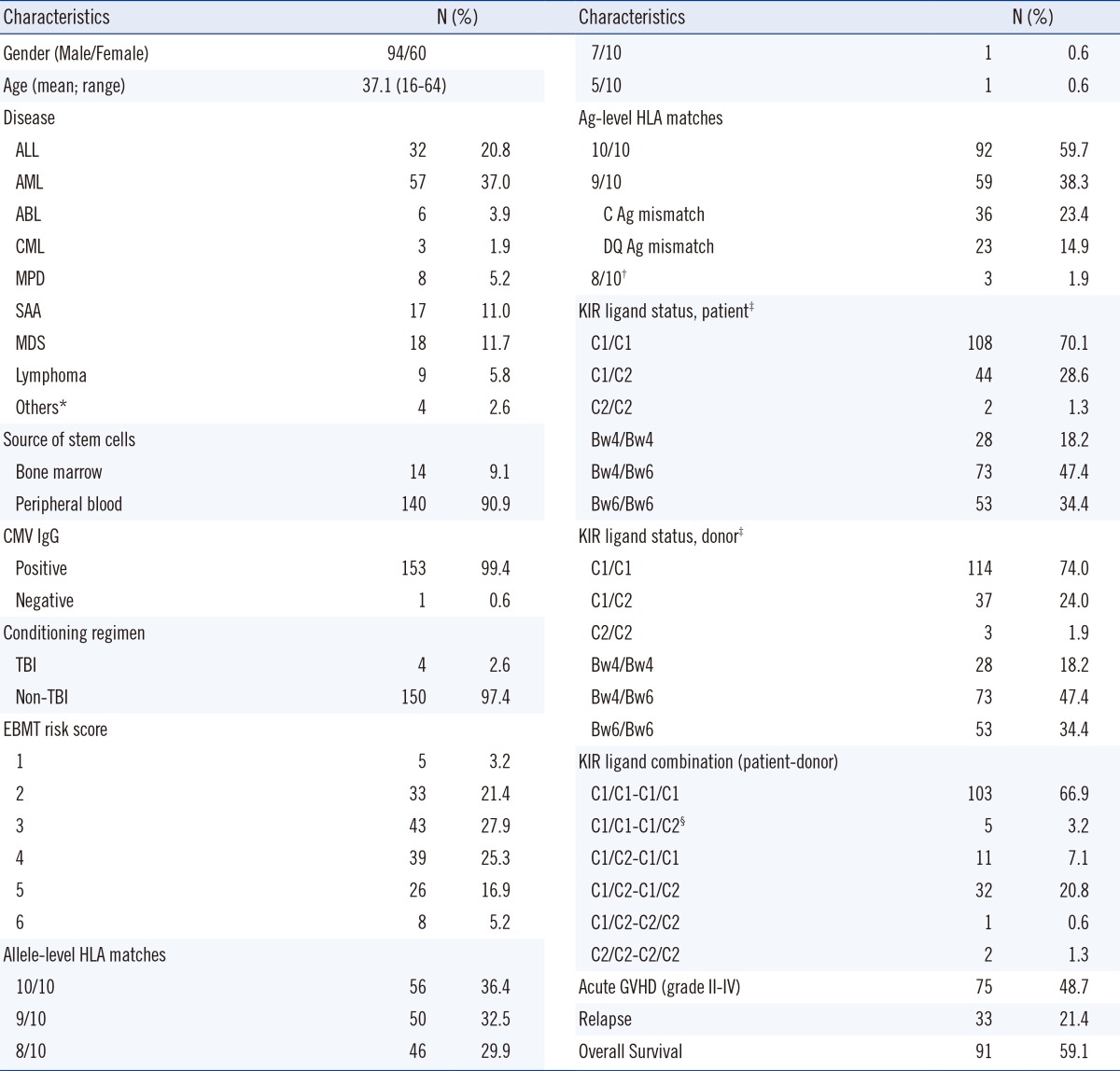
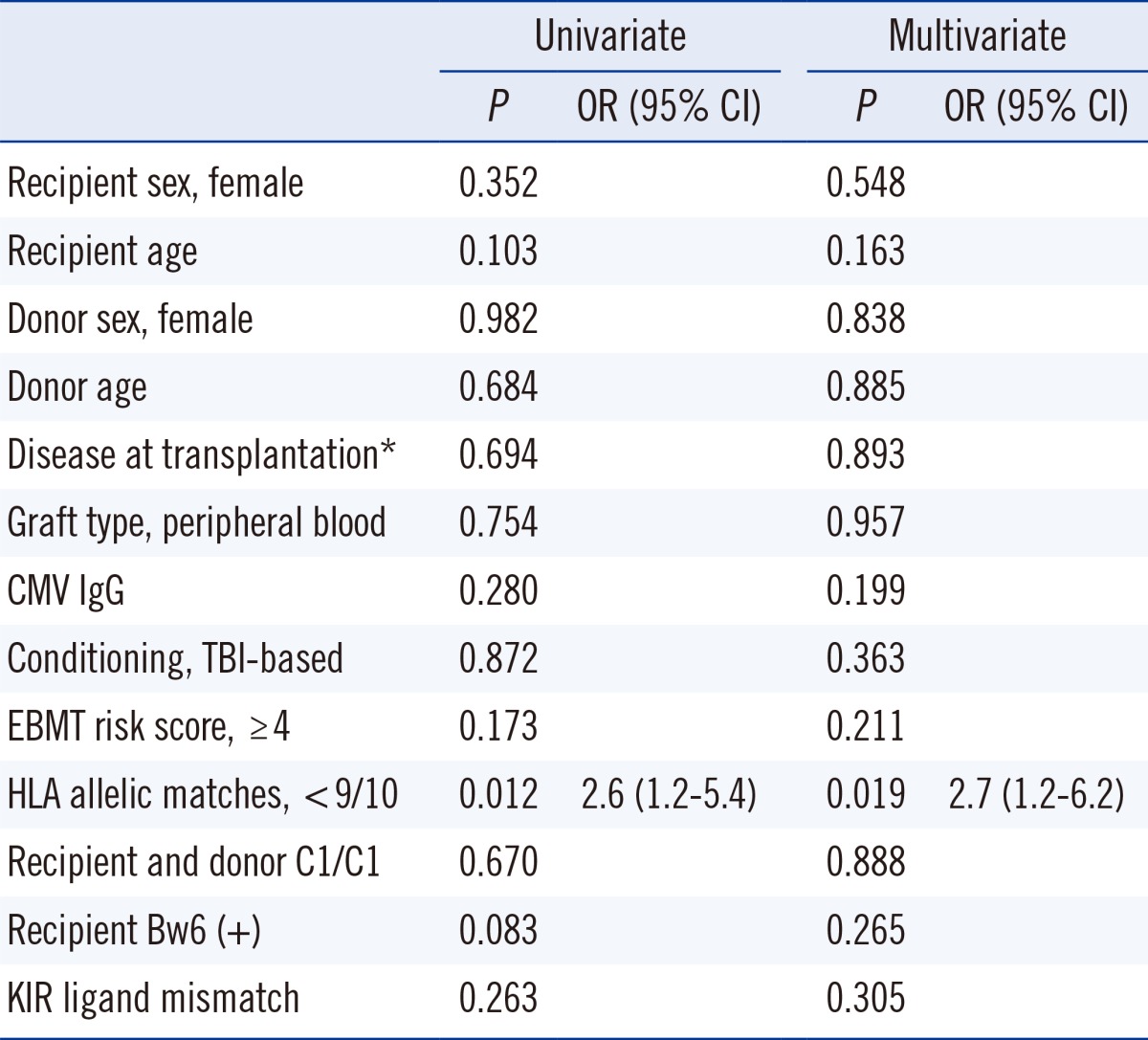
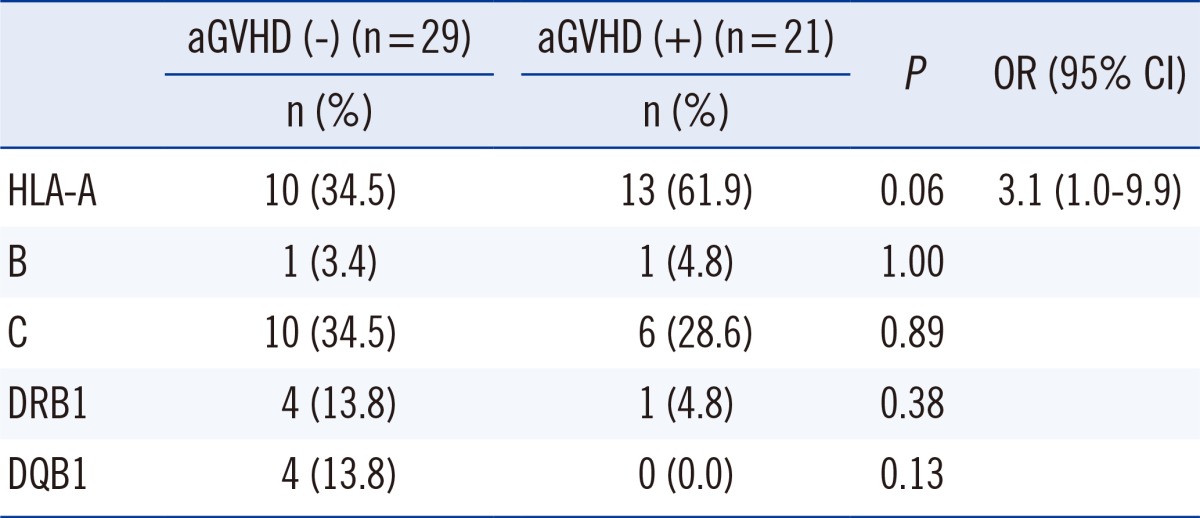




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download