Abstract
Background
Human neutrophil antigens (HNAs) are involved in autoimmune and alloimmune neutropenia and transfusion-related acute lung injury. The HNA-1 system is important in immunogenetics, and allele frequencies have been described in different populations. This study investigated the frequency of FCGR3B alleles encoding HNA-1a, HNA-1b, and HNA-1c among Thai blood donors and compared these frequencies with those previously reported for other populations.
Methods
Eight hundred DNA samples obtained from unrelated healthy blood donors at the National Blood Centre, Thai Red Cross Society, Bangkok, and the Blood Bank, Faculty of Medicine, Chiang Mai University, Chiang Mai, Thailand, were included. Samples were simultaneously typed for each FCGR3B allele using an in-house polymerase chain reaction with sequence-specific primer (PCR-SSP) technique.
Results
The frequencies of FCGR3B*1, FCGR3B*2, and FCGR3B*3 alleles in central Thai blood donors were 0.548, 0.452, and 0.004, respectively; only FCGR3B*1 and FCGR3B*2 alleles were found in northern Thai blood donors (0.68 and 0.32, respectively). Compared with other Asian populations, central Thais had higher frequencies of the FCGR3B*2 allele (P<0.001), while the frequencies of the FCGR3B*1 and FCGR3B*2 alleles in northern Thais were similar to those previously reported in Taiwanese and Japanese populations. In contrast, the frequencies of the FCGR3B*1 and FCGR3B*2 alleles in the northern Thai population were statistically different from those observed in central Thai, Korean, German, and Turkish populations.
In Thailand, the demographic Thai populations include the central, northeastern, northern, and southern Thais. The northern Thai population is one of the major subgroups of ethnic Thais and has close cultural and historic connections with the populations of southern Chinese, Myanmar and Laos. A previous study on the HLA system in 5 ethnic Thai populations recruited from central, northeastern, and northern Thailand demonstrated that the HLA-DRB1*15:02 allele appeared to be dominant in Thais; however, the HLA-DRB1*15:01 and the HLA-DRB1*09:01 alleles were relatively more common in northern Thais than other ethnic Thais [1]. This evidence supports the hypothesis that the heterogeneous southern populations of East Asia are genetically derived from the population of Southeast Asia [2, 3].
Human neutrophil antigens (HNA) are polymorphic structures located in the membranes of neutrophils [4]. Currently, 5 HNA systems (HNA-1 to HNA-5), have been characterized [5, 6]. In the HNA-1 system, 3 antigens, HNA-1a, HNA-1b and HNA-1c, are located on the neutrophil low-affinity Fc-γ-IIIb receptor (CD 16) glycoprotein [7]. Because Fc-γ-IIIb is the most immunogenic glycoprotein on the neutrophil membrane, antibodies to these antigens are frequently implicated in alloimmune and autoimmune neutropenia and transfusion-related acute lung injury (TRALI) [8-12].
The Fc-γ-IIIb receptor is encoded by the FCGR3B gene and occurs in 3 polymorphic forms representing the human neutrophil antigens, that is, HNA-1a, HNA-1b, and HNA-1c, which are encoded by the FCGR3B*1, FCGR3B*2, and FCGR3B*3 alleles, respectively [13]. In addition to their clinical significance, the FCGR3B allele frequencies could be used as an additional marker in population studies [14-16]. Moreover, the HNA genetic system of Thai populations has not yet been adequately studied. Therefore, this study aimed to determine the FCR3B*1, FCGR3B*2, and FCGR3B*3 allele frequencies encoding HNA-1a, HNA-1b, and HNA-1c, respectively, in Thai blood donors by using a PCR sequence-specific primer (PCR-SSP) technique to compare the allele frequencies with those previously reported in other populations.
Peripheral venous blood was collected in EDTA-anticoagulated vacutainer from 800 unrelated healthy Thai blood donors. Five hundred samples, that is, 300 samples from our previous study [17] and an additional 200 samples, were from the National Blood Centre, Thai Red Cross Society, Bangkok. In addition, 300 samples were from the Blood Bank, Faculty of Medicine, Chiang Mai University, Chiang Mai, Thailand. Written informed consent was obtained from each subject. This study was approved by the Committee on Human Rights Related to Research Involving Human Subjects, Thammasat University, Pathumtani, Thailand. Genomic DNA was extracted from peripheral blood samples using the Genomic DNA extraction kit (REAL Genomics, RBCBioscience, Taipei, Taiwan) and by a salting out method, as previously described [18], and was then stored at -20℃ until use for genotyping.
Known HNA-1a, HNA-1b, and HNA-1c DNA samples were provided by Dr. Núria Nogués, Laboratori d'Immunohematologia, Banc de Sang i Teixits, Passeig Taulat, Barcelona, Spain. A PCR-SSP technique was performed as previously described [9] with some modifications. The sequences of the primers used for HNA genotyping are shown in Table 1. The specific primers were similar to those previously described [4, 9]. Briefly, 2 µL of genomic DNA (50 ng/µL) was amplified in a total volume of 10 µL with 0.5 µM of HNA-1a forward (F) and reverse (R) primers for FCGR3B*1 genotyping, 0.5 µM of HNA-1b (F) and (R) primers for FCGR3B*2 genotyping, and 0.5 µM of HNA-1c (F) and (R) primers for FCGR3B*3 genotyping. The human growth hormone (HGH) gene was coamplifed using 0.125 µM HGH (F) primer and 0.125 µM HGH (R) primer and was used as an internal control. PCR was performed with 5 µL of DreamTaq DNA polymerase (Thermo Fisher Scientific Inc., Glen Burnie, MD, USA) consisting of 2X DreamTaq Green buffer, 0.4 mM of each of the dNTPs, and 0.4 mM MgCl2 in a G-STORM GS1 thermal cycler (Gene Technologies Ltd., Somerton, UK). The cycling parameters for the PCR program were as follows: 1 cycle of 300 sec at 95℃; 30 cycles each of 30 sec at 95℃, 60 sec at 57℃, and 30 sec at 72℃; and final extension for 5 min at 72℃. The product was then kept at 4℃ until use for analysis. After amplification, the PCR products were analyzed on a 2.0% agarose gel using 1X Tris borate ethylenediaminetetraacetate (TBE) buffer containing SYBR Green I nucleic acid gel stain (Invitrogen, Grand Island, NY, USA) and were visualized under ultraviolet (UV) illumination (Fig. 1).
Gene frequencies were calculated by gene counting as previously described [4, 19]. The Chi-square test was used to evaluate whether the observed genotype frequencies were in agreement with the expected ones under Hardy-Weinberg equilibrium. Genotype and allele frequencies of FCGR3B*1, FCGR3B*2, and FCGR3B*3 were compared between different populations by using the Chi-square test. P values of less than 0.05 were considered statistically significant.
The FCGR3B genotypes and allele frequencies among different populations are shown in Table 2. For the central Thai blood donors, the FCGR3B*1 allele was most frequently detected (0.548 [548/1,000]), followed by FCGR3B*2 (0.452 [452/1,000]), with FCGR3B*3 being the least frequent (0.004 [4/1,000]). FCGR3B*1/FCGR3B*2 was the most frequent genotype (321/500), followed by FCGR3B*1/FCGR3B*1 (112/500) and FCGR3B*2/FCGR3B*2 (63/500). On the other hand, only 2 alleles, FCGR3B*1 and FCGR3B*2, were found in the northern Thai population, with allele frequencies of 0.677 (406/600) and 0.323 (194/600), respectively. In this population, FCGR3B*1/FCGR3B*1 was the most frequent genotype (140/300), followed by FCGR3B*1/FCGR3B*2 (126/300) and FCGR3B*2/FCGR3B*2 (34/300). We did not find any HNA-1 null subjects among the 800 samples investigated in the present study. Moreover, the FCGR3B locus was in Hardy-Weinberg equilibrium in Thai populations. The genotype frequency distribution of the central Thai population was also compared with that of the northern Thai population and other studies previously reported [9, 14, 16, 20, 21]. The FCGR3B*1 and FCGR3B*2 allele frequencies of the northern Thais did not differ from those of the Taiwanese and Japanese populations [9, 14]. Interestingly, the FCGR3B*1 and FCGR3B*2 allele frequencies were significantly different from those observed in the central Thai, German, Turkish, Tunisian, and Korean populations (P<0.01) [16, 20, 21].
In this study, the FCGR3B alleles encoding the HNA-1a, -1b, and -1c antigens were analyzed by the in-house PCR-SSP technique, because the commercial kit for HNA-1a, HNA-1b, and HNA-1c genotyping is expensive and is not available in Thailand. Although the PCR-SSP technique is not suitable for high-throughput genotyping, it is widely used in routine laboratory examinations for both HLA and HNA genotyping because of its simplicity. Moreover, in order to simultaneously read the HNA-1 genotyping results, the same PCR conditions were used for each FCGR3B allele detection, which makes this technique less time consuming than the PCR-SSP technique previously reported [9]. Additionally, the cost of this in-house PCR-SSP technique is approximately only 1 US dollar per test.
It was found that the FCGR3B*1 allele was the most common allele in 800 Thai blood donors. This finding confirms that the FCGR3B*1 allele is more frequent than the FCGR3B*2 allele in Asian populations, Afro-Brazilians, and Native Americans [4, 6, 7, 9, 13, 14]. On the other hand, the FCGR3B*1 and FCGR3B*2 allele frequencies in the northern Thai population were significantly different from those in the central Thai and Korean populations [20]. The FCGR3B*3 allele, which is considered to be a rare allele in Asian populations [9], was found only in central Thai blood donors, similar to the findings of a previous study [17]. This allele was present in 9.09% of Tunisian blood donors and only in 5% of European populations [16].
HNA-1a individuals with the FCGR3B*1/FCGR3B*1 genotype are reported to be more susceptible to idiopathic pulmonary fibrosis, whereas HNA-1b individuals with the FCGR3B*2/FCGR3B*2 genotype have a higher susceptibility to periodontal diseases and chronic periodontitis because of differing reactivity to opsonised bacteria [22]. Therefore, diseases associated with HNA-1 antigens found in the northern Thai population are different from those for the central Thai population. Further studies are required on the association of HNA-1 and disease susceptibility. Additionally, a higher frequency of the FCGR3B*1 allele and a lower frequency of the FCGR3B*2 allele were found in northern Thai blood donors compared with the central Thai population, suggesting that northern Thais would be more susceptible to HNA-1a alloimmunization than the central Thais, similar to other Asian populations [14, 20].
Our results demonstrate that the FCGR3B*1 and FCGR3B*2 allele frequencies significantly differ between the central and northern Thai populations. However, the allele frequencies in the northern Thai population are closely related to those for the Taiwanese and Japanese populations. In addition, further studies on HNA-3, HNA-4, and HNA-5 gene frequencies would be beneficial in order to develop a database of HNA characteristics in Thai populations.
Figures and Tables
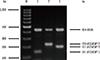
Fig. 1
PCR sequence-specific primer analysis of the FCGR3B*1-, FCGR3B*2-, and FCGR3B*3-positive individuals. The 141-bp, 219-bp, and 191-bp bands represent the specific PCR products and the 434-bp HGH bands are the internal controls. PCR products were analyzed on a 2.0% agarose gel stained with ethidium bromide. The DNA size standard (100-bp ladder marker; Fermentas, Carlsbad, CA, USA) is shown in lane M.
Acknowledgements
This work is supported by the National Research University Project of Thailand, Office of Higher Education Commission. We thank Dr. Nuria Nogues, Laboratori d'Immunohematologia, Banc de Sang i Teixits, Passeig Taulat, Barcelona, Spain, for providing the DNA controls.
References
1. Stephens HA, Chandanayingyong D, Kunachiwa W, Sirikong M, Longta K, Maneemaroj R, et al. A comparison of molecular HLA-DR and DQ allele profiles forming DR51-, DR52-, and DR53-related haplotypes in five ethnic Thai populations from mainland southeast Asia. Hum Immunol. 2000; 61:1039–1047.

2. Chu JY, Huang W, Kuang SQ, Wang JM, Xu JJ, Chu ZT, et al. Genetic relationship of populations in China. Proc Natl Acad Sci USA. 1998; 95:11763–11768.

4. de La Vega Elena CD, Nogués N, Fernándes Montoya A, Oyonarte S, Solis E, Muñiz-Dias E. HNA-1a, HNA-1b and HNA-1c gene frequencies in Argentineans. Tissue Antigens. 2008; 71:475–477.

6. Xia W, Bayat B, Sachs U, Chen Y, Shao Y, Xu X, et al. The frequencies of human neutrophil alloantigens in the Chinese Han population of Guangzhou. Transfusion. 2011; 51:1271–1277.

7. Chu CC, Lee HL, Chu TW, Lin M. The use of genotyping to predict the phenotypes of human platelet antigens 1 through 5 and of neutrophil antigens in Taiwan. Transfusion. 2001; 41:1553–1558.

8. Bux J, Stein EL, Bierling P, Fromont P, Clay M, Stroncek D, et al. Characterization of a new alloantigen (SH) on the human neutrophil Fc gamma receptor IIIb. Blood. 1997; 89:1027–1034.
9. Kissel K, Hofmann C, Gittinger FS, Daniels G, Bux J. HNA-1a, HNA-1b, and HNA-1c (NA1, NA2, SH) frequencies in African and American Blacks and in Chinese. Tissue Antigens. 2000; 56:143–148.

10. Bux J, Jung KD, Kauth T, Mueller-Eckhardt C. Serological and clinical aspects of granulocyte antibodies leading to alloimmune neonatal neotropenia. Transfus Med. 1992; 2:143–149.
11. Lalezari P, Jiang AF, Yegen L, Santorineou M. Chronic autoimmune neutropenia due to anti-NA2 antibody. N Engl J Med. 1975; 293:744–747.

12. Yomtovian R, Kline W, Press C, Clay M, Engman H, Hammerschmidt D, et al. Severe pulmonary hypersensitivity associated with passive transfusion of a neutrophil-specific antibody. Lancet. 1984; 1:244–246.

13. Covas DT, Kashima S, Gerrreiro JF, dos Santos SE, Zago MA. Variation in the FcγR3B gene among distinct Brazilian populations. Tissue Antigens. 2005; 65:178–182.
14. Fujiwara K, Watanabe Y, Mitsunaga S, Oka T, Yamane A, Akaza T, et al. Determination of granulocyte-specific antigens on neutrophil FcA receptor IIIb by PCR-preferential homoduplex formation assay, and gene frequencies in the Japanese population. Vox Sang. 1999; 77:218–222.
15. Kuwano ST, Bordin JO, Chiba AK, Mello AB, Figueiredo MS, Vieira-Filho JP, et al. Allelic polymorphisms of human fcgamma receptor IIa and Fcgamma receptor IIIb among distinct groups in Brazil. Transfusion. 2000; 40:1388–1392.

16. Hassine MO, Ennafaa H, Kalai S, Kibech R, Sellami MH, Bouzid L, et al. FCGR3B allele frequencies in Tunisians of sub-Saharan origin. Transfus Clin Biol. 2012; 19:60–63.
17. Changsri K, Tobunluepop P, Songthammawat D, Apornsuwan T, Kaset C, Nathalang O. Human neutrophil alloantigen genotype frequencies in Thai blood donors. Blood Transfus. 2013; 01. 23. 1–6. doi: 10.2450/2013.0161-12 [Epub ahead of print].
18. Miller AS, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA for human nucleated cells. Nucleic Acids Res. 1988; 16:1215.
19. Steffensen R, Gülen T, Varming K, Jersild C. FcγRIIIB polymorphism: evidence that NA1/NA2 and SH are located in two closely linked loci and that the SH allele is linked to the NA1 allele in the Danish population. Transfusion. 1999; 39:593–598.
20. Seo DH, Park SS, Han KS. Genotype analysis of granulocyte-specific antigens in Koreans. Korean J Clin Pathol. 1997; 17:1144–1149.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download