Dear Editor
The new 2008 WHO classification of hemato-oncological diseases includes a new category for recurrent chromosomal abnormalities [1]. Recurrent gene rearrangements such as RUNX1/RUNX1T1 (formerly AML1/ETO), CBFB/MYH11, or PML/RARA, provide additional information for the diagnosis of AML, regardless of the blast count. Chromosomal abnormalities and gene mutations are the most influential and/or prominent factors used for determining prognosis. Detection of recurrent genetic abnormalities has become important for the diagnosis, determination of prognosis, and detection of minimal residual disease. Many translocations can be detected through conventional cytogenetics, FISH, and reverse transcriptase-PCR (RT-PCR). Among these, RT-PCR is the most sensitive method and can also detect cryptic gene rearrangements that remain undetected by conventional cytogenetics [2]. Single RT-PCR has been the most commonly used method to detect gene rearrangements in Korea. However, this method is limited to the detection of only certain types of translocations. In the 2000s, multiplex RT-PCR became widely adopted in Korea, enabling rapid detection of a broader range of gene rearrangements.
In this study, we compared the diagnostic utility of multiplex RT-PCR to single RT-PCR, and then compared both of them with conventional cytogenetics. Although a direct comparison was not available because of different sampling periods, we investigated the difference between single RT-PCR and conventional cytogenetics to provide a more robust assessment of the diagnostic utility of multiplex RT-PCR.
Among patients who visited our institution from January 2005 to June 2012, we identified individuals who were diagnosed with hematologic malignancies. Single RT-PCR was used for the diagnosis of patients from January 2005 to May 2008, and multiplex RT-PCR from June 2008 to June 2012. Patients were considered to have a hematologic malignancy when the results indicated AML, ALL, mixed phenotype acute leukemia (MPAL), and CML. Cytogenetic analysis and FISH analysis were performed according to the manufacturer's instructions. Single RT-PCR was performed using 3 primers for PML/RARA, BCR/ABL1, and RUNX1/RUNX1T1. Multiplex RT-PCR was conducted by the HemaVision kit (DNA Technology, Aarhus, Denmark).
Characteristics of patients diagnosed with hematologic malignancies by single RT-PCR and multiplex RT-PCR are shown in Table 1. There were 65 patients who were diagnosed using single RT-PCR. Two of these patients who were initially diagnosed with AML and ALL, respectively, experienced relapse and were subsequently given the same diagnosis. In terms of diagnostic outcomes, AML was the most frequently diagnosed hematologic malignancy (36 cases). Further, there were 14 cases of ALL (13 B-cell [B-ALL] and 1 T-cell [T-ALL]), 3 cases of MPAL, 1 case of unclassified acute leukemia due to unavailability of a bone marrow biopsy specimen (peripheral blood test was subsequently conducted), and 11 cases of CML. Using multiplex RT-PCR, we detected hematologic malignancies in 81 patients. Among them, 77 were found to be ill on initial diagnosis and 4 experienced relapses. Among the patients who experienced relapse, 2 with AML and ALL, respectively, received the same diagnosis before and after relapse. The remaining 2 patients were diagnosed with AML at their second relapse. Similarly, AML was found to be the most frequently diagnosed hematologic malignancy (53 cases) using multiplex RT-PCR. Further, there were 16 cases of ALL (15 B-ALL and 1 T-ALL), 1 case of MPAL, 1 case of unclassified acute leukemia due to unavailability of a bone marrow biopsy specimen (peripheral blood test was subsequently conducted), and 10 cases of CML. Translocations were detected in 40% (26/65) of the patients by single RT-PCR and 51.9% (42/81) of the patients by multiplex RT-PCR. For statistical analysis, chi-square test was conducted using SPSS version 19.0 (SPSS Inc., Chicago, IL, USA). However, no statistically significant difference was observed in the positivity rate between the 2 groups of patients (P=0.154).
Single RT-PCR detected 18 cases of BCR/ABL1 gene rearrangement, 4 cases of RUNX1/RUNX1T1 gene rearrangement, and 4 cases of PML/RARA gene rearrangement. In contrast, 22 cases of BCR/ABL1 gene rearrangement, 7 cases of PML/RARA gene rearrangement, 4 cases of RUNX1/RUNX1T1 gene rearrangement, 3 cases of CBFB/MYH11 gene rearrangement, 2 cases of ETV6/RUNX1 gene rearrangement, 1 case of MLL/ELL gene rearrangement, 1 case of MLL/MLLT3 gene rearrangement, 1 case of RUNX1/MDS1/EVI1 gene rearrangement, and 1 case of NPM1/MLF1 gene rearrangement were detected by multiplex RT-PCR analysis. While single RT-PCR was only able to detect 3 frequently reported gene rearrangements (RUNX1/RUNX1T1, PML/RARA, and BCR/ABL1), multiplex RT-PCR identified several other types of gene rearrangement, including a rare NPM1/MLF1 rearrangement. To the best of our knowledge, this is the first time this type of rearrangement has been detected and reported in Korea [3]. Apart from the 28 different translocations it can identify, the HemaVision kit (DNA Technology) can also effectively detect more than 80 breakpoints. Because the HemaVision kit can easily detect atypical breakpoints in BCR/ABL1, we were able to identify a unique e8a2 fusion transcript that has not yet been reported in B-ALL cases [4].
Single RT-PCR produced negative results for 3 cases that were subsequently tested positive after cytogenetic analysis (Table 2); 2 of these cases showed translocations at inv(16) and t(4;11). If these patients had undergone testing using multiplex RT-PCR, CBFB/MYH11 or MLL/AFF1 gene rearrangements may have been detected. Considering how many clinically important hematologic malignancy-related fusion genes and translocations are being reported [5], and because chromosome abnormalities affect the prognosis of diseases other than leukemogenesis, prompt detection of genetic changes is imperative.
In this study, ETV6/RUNX1 gene rearrangements were detected in 2 cases of B-ALL with normal karyotypes using multiplex RT-PCR (Table 3). FISH results showed ETV6/RUNX1 gene rearrangements in 55.8% and 80.7% of specimens analyzed. ETV6/RUNX1 is known to be the most common molecular genetic abnormality and is found in about 20-25% of pediatric cases of B-ALL [6]. However, as a cryptic translocation, ETV6/RUNX1 is difficult to detect with conventional cytogenetics and therefore requires RT-PCR or FISH as a parallel test [7]. The study also found a case of MLL/ELL gene rearrangements with a normal karyotype (Table 3). Although R-banding can identify where chromosome 11 is enlarged, a MLL/ELL abnormality can be mistaken for a normal karyotype under G-banding [8]. Therefore, RT-PCR or FISH would be necessary to confirm cytogenetic analysis results. Multiplex RT-PCR by HemaVision (DNA Technology) is limited by the fact that it can only detect about 10 out of 70 known MLL partner gene types [9-11]. In order to enhance the sensitivity of gene abnormality detection in patients with hematologic malignancies but normal karyotypes, multiplex RT-PCR kits should be further developed to detect additional types of MLL gene rearrangements.
This study is limited in that a small number of hematologic malignancy cases were included; moreover, only a few of the patients analyzed had positive PCR results. Further, this was a retrospective analysis; here, a comparison was made between different individuals who visited our institution during different periods, and the methods of single RT-PCR or multiplex RT-PCR were used separately.
In conclusion, a combination of multiplex RT-PCR and conventional cytogenetics should be considered the routine procedure for the diagnosis of hematologic malignancies. Multiplex RT-PCR panels should be developed to increase diagnostic capability and treatment effectiveness for hematologic malignancies.
References
1. Swerdlow SH, Campo E, editors. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th ed. Lyon: IARC Press;2008.
2. King RL, Naghashpour M, Watt CD, Morrissette JJ, Bagg A. A comparative analysis of molecular genetic and conventional cytogenetic detection of diagnostically important translocations in more than 400 cases of acute leukemia, highlighting the frequency of false-negative conventional cytogenetics. Am J Clin Pathol. 2011; 135:921–928. PMID: 21571965.

3. Lim G, Choi JR, Kim MJ, Kim SY, Lee HJ, Suh JT, et al. Detection of t(3;5) and NPM1/MLF1 rearrangement in an elderly patient with acute myeloid leukemia: clinical and laboratory study with review of the literature. Cancer Genet Cytogenet. 2010; 199:101–109. PMID: 20471513.

4. Kim MJ, Yoon HJ, Park TS. The e8a2 fusion transcript in B lymphoblastic leukemia with BCR-ABL1 rearrangement. Korean J Hematol. 2012; 47:161. PMID: 23071469.
5. Mitelman F, Mertens F, Johansson B. Prevalence estimates of recurrent balanced cytogenetic aberrations and gene fusions in unselected patients with neoplastic disorders. Genes Chromosomes Cancer. 2005; 43:350–366. PMID: 15880352.

6. Jamil A, Theil KS, Kahwash S, Ruymann FB, Klopfenstein KJ. TEL/AML-1 fusion gene. its frequency and prognostic significance in childhood acute lymphoblastic leukemia. Cancer Genet Cytogenet. 2000; 122:73–78. PMID: 11106814.
7. Kim SR, Kim HJ, Kim SH. Clinical utility of fluorescence in-situ hybridization profile test in detecting genetic aberrations in acute leukemia. Korean J Lab Med. 2009; 29:371–378. PMID: 19893343.

8. Huret JL. t(11;19)(q23;p13.1). Atlas Genet Cytogenet Oncol Haematol. 1997; 1:102–103. http://AtlasGeneticsOncology.org/Anomalies/t1119ELL.html.
9. Park TS, Lee SG, Song J, Lee KA, Kim J, Choi JR, et al. CASP8AP2 is a novel partner gene of MLL rearrangement with t(6;11) (q15;q23) in acute myeloid leukemia. Cancer Genet Cytogenet. 2009; 195:94–95. PMID: 19837277.
10. Kim MJ, Choi JR, Suh JT, Lee HJ, Lee WI, Park TS. Diagnostic standardization of leukemia fusion gene detection system using multiplex reverse transcriptase-polymerase chain reaction in Korea. J Korean Med Sci. 2011; 26:1399–1400. PMID: 22022200.
11. Yang JJ, Marschalek R, Meyer C, Park TS. Diagnostic usefulness of genomic breakpoint analysis of various gene rearrangements in acute leukemias: a perspective of long distance- or long distance inverse-PCR-based approaches. Ann Lab Med. 2012; 32:316–318. PMID: 22779077.

12. Shaffer LG, Slovak ML, editors. ISCN (2009): an international system for human cytogenetic nomenclature. Basel: S. Karger;2009.
Table 1
Characteristics of patients diagnosed with hematologic malignancies by single and multiplex RT-PCR analysis
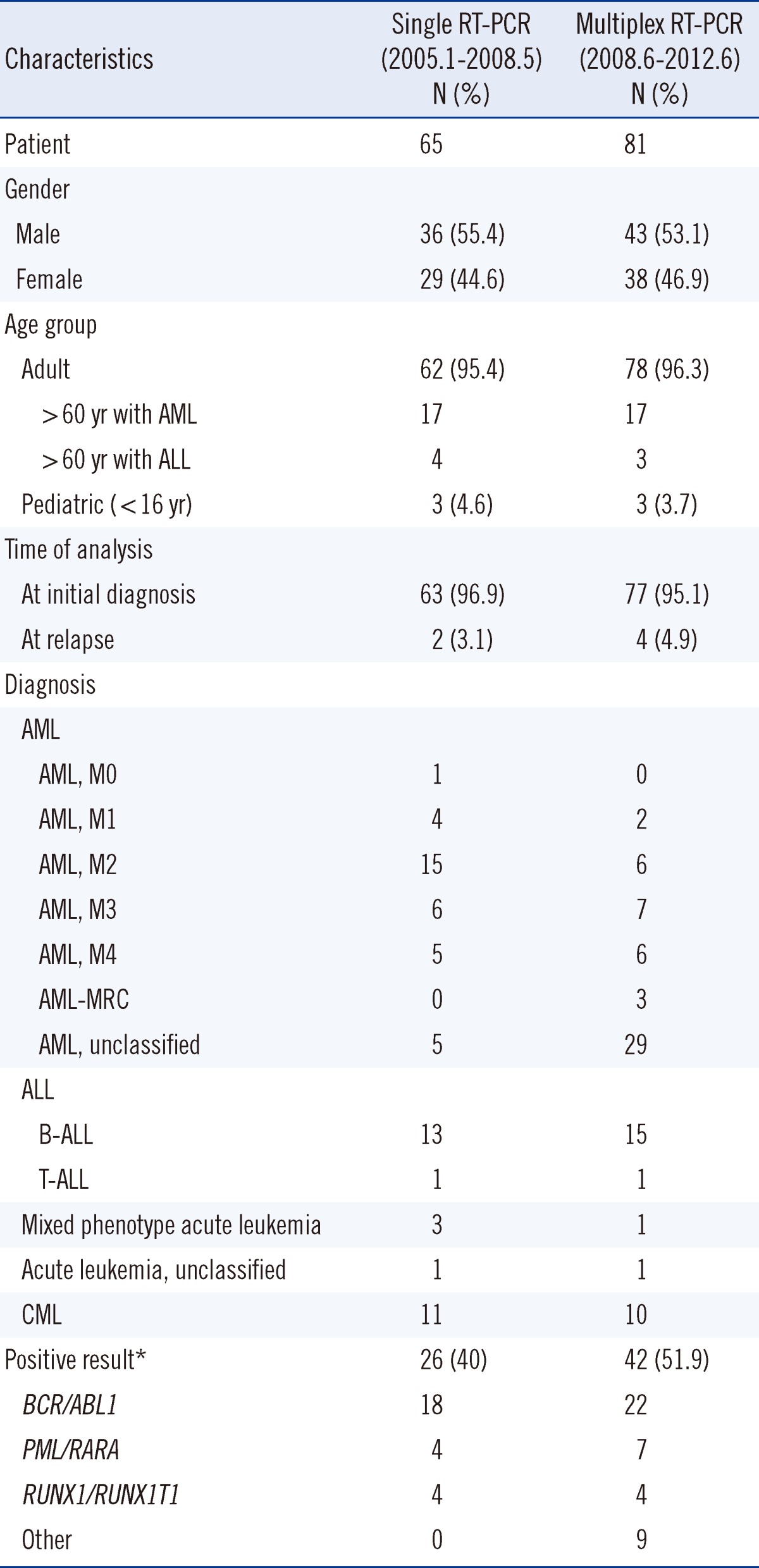
Table 3
Discrepancy between cytogenic analysis and multiplex RT-PCR results

*ETV6/RUNX1 is the new abbreviation for TEL/AML1, as provided by the HUGO Gene Nomenclature Committee [12].
Abbreviations: RT, reverse transcriptase; F, female; M, male.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download