Abstract
Background
The aims of this study were to understand the molecular epidemiology of integron-associated gene cassettes in Acinetobacter baumannii across four hospitals in northern Taiwan and to clarify the relationship between the presence of integrons and antibiotic-resistant phenotypes.
Methods
Sixty-five A. baumannii isolates, collected from the patients of four regional hospitals in northern Taiwan in 2009, were tested for the presence of integrons and their associated gene cassettes. The susceptibility difference between integron-positive and integron-negative A. baumannii strains was analyzed. Antibiotic-resistant phenotypes among A. baumannii with different types of gene cassette array combinations were also compared.
Results
Around 72% of the A. baumannii isolates carried class 1 integrase genes. Despite this, only three gene cassette arrays were found in the integrons. Integron-positive strains were significantly more resistant to all the tested antibiotics than the integrase-negative strains. All the four types of A. baumannii with different gene cassette array combinations were multidrug-resistant in nature. Gene cassette array aacA4-catB8-aadA1 existed in all the integron-positive A. baumannii isolates. Repetitive-sequence-based PCR (rep-PCR) results revealed the prevalence of one major cluster of imipenem-resistant A. baumannii strains (84%) in the four regional hospitals.
Over the past two decades, multidrug-resistant Acinetobacter baumannii has been globally recognized as an important cause of nosocomial infections [1]. Both uncanny environmental adaptation and rapid acquisition of resistant genes contribute to the ability of A. baumannii to be such a successful pathogen [2]. The spread of these resistant genes is mainly through horizontal transfer of plasmids, transposons, or integrons that carry clusters of genes resistant to several antibiotic families [3]. Of these antibiotic-resistance mechanisms, integrons are considered unique for their capacity to cluster and express resistance genes [4]. At present, five classes of mobile integrons are known to have a role in the dissemination of antibiotic-resistance genes. Class 1 integrons are most commonly found in the clinical isolates of gram-negative bacteria and more than hundred accessory genes are known to confer resistance. By contrast, only six different resistance cassettes have been found that are associated with class 2 integrons [5]. Previous studies showed that class 1 integrons and the pool of associated gene cassettes are major contributors to the multidrug resistance of A. baumannii [6, 7]. As a result, integrons are regarded as a useful marker for the epidemiological study of A. baumannii [8].
The aims of this study were to understand the molecular epidemiology of integron-associated gene cassettes in A. baumannii among the four hospitals in northern Taiwan and to clarify the relationship between the presence of integrons and antibiotic-resistant phenotypes.
This study was conducted in Tao-Yuan, Hsin-Chu, Chut-Tung, and Miao-Li General Hospitals, Department of Health (DOH), Executive Yuan, Taiwan between June and December in 2009. Ten imipenem-resistant and ten imipenem-susceptible Acinetobacter species were consecutively collected in these four regional hospitals. Each isolate was sampled from different patients. Organisms were classified as belonging to the genus Acinetobacter by using the Vitek system (Biomerieux Vitek, Inc., Hazelwood, MO, USA). Identification of these isolates as A. baumannii was performed by one-tube multiplex PCR, based on the method of Chen et al. [9].
Susceptibilities to antimicrobial agents were determined by the disk diffusion method, in accordance with the guidelines of the Clinical and Laboratory Standards Institute [10]. The agents tested included ampicillin/sulbactam, piperacillin/tazobactam, gentamicin, amikacin, ciprofloxacin, levofloxacin, trimethoprim/sulfamethoxazole, ceftazidime, and imipenem.
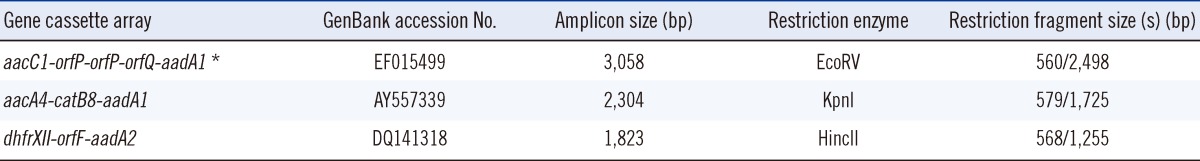
Detection of class 1 and 2 integrase genes (intI1 and intI2) and integron gene cassettes was performed by PCR, as described previously [11]. Integron gene cassettes were detected using 5' CS and 3' CS primers. The target gene fragments were isolated with the Gel Elution Kit, FavorPrep (Favorgen Biotech Corporation, Ping-Tung, Taiwan), and the yT&A Cloning Kit from Yeastern Biotech Co., Ltd. (Taipei, Taiwan). Confirmation of the gene cassette sizes was performed by restriction fragment length polymorphism (RFLP) with the help of NEBcutter V2.0 (New England Biolab Inc. Ipswich, MA, USA) to verify the enzyme cutting sites [12]. The expected sizes of amplified DNA fragments confirmed the identity of the gene cassettes. The integron gene cassette amplicons were then sequenced and compared to those registered in the National Center for Biotechnology Information (NCBI) database. Amplification fragments that were not of the expected sizes were subjected to further sequencing.
To clarify the relationship between the genotypes of imipenem-resistant A. baumannii and the distribution of integrons with gene cassettes, repetitive-sequence-based PCR (rep-PCR) was performed for DNA fingerprinting as described by Misbah et al. [13]. REP-1 (NNN NCG NCG NCA TCN GGC) and REP-2 (NCG NCT TAT CNG GCC TAC) primers were used for DNA amplification. The PCR conditions for amplification were as follows: initial denaturation at 94℃ for 5 min, followed by 30 cycles of denaturation at 90℃ for 30 sec, primer annealing at 32℃ for 1 min, and primer extension at 65℃ for 8 min. Amplification products were resolved by electrophoresis at 50 V for 2.5 hr on 1.8% agarose gels. After staining with ethidium bromide, each gel was visualized under UV light. The rep-PCR profiles of the A. baumannii were analyzed with Numerical Taxonomy and Multivariate Analysis System version 2.0 (Applied Biostatistics Inc., Setauket, NY, USA). Cluster analysis for rep-PCR DNA fingerprints was performed by construction of a dendrogram, which was calculated by using the band-based dice coefficient method. A similarity matrix was generated and subsequently clustered using the unweighted pair group method with arithmetic means.
The susceptibility difference between integron-positive and integron-negative A. baumannii strains was analyzed by chi-square or Fisher's exact test, as appropriate. The differences between two groups of isolates were considered significant at P<0.05. Data entry and analyses were performed using the Statistical Package for the Social Sciences (SPSS) software version 15.0 (SPSS Inc., Chicago, IL, USA).
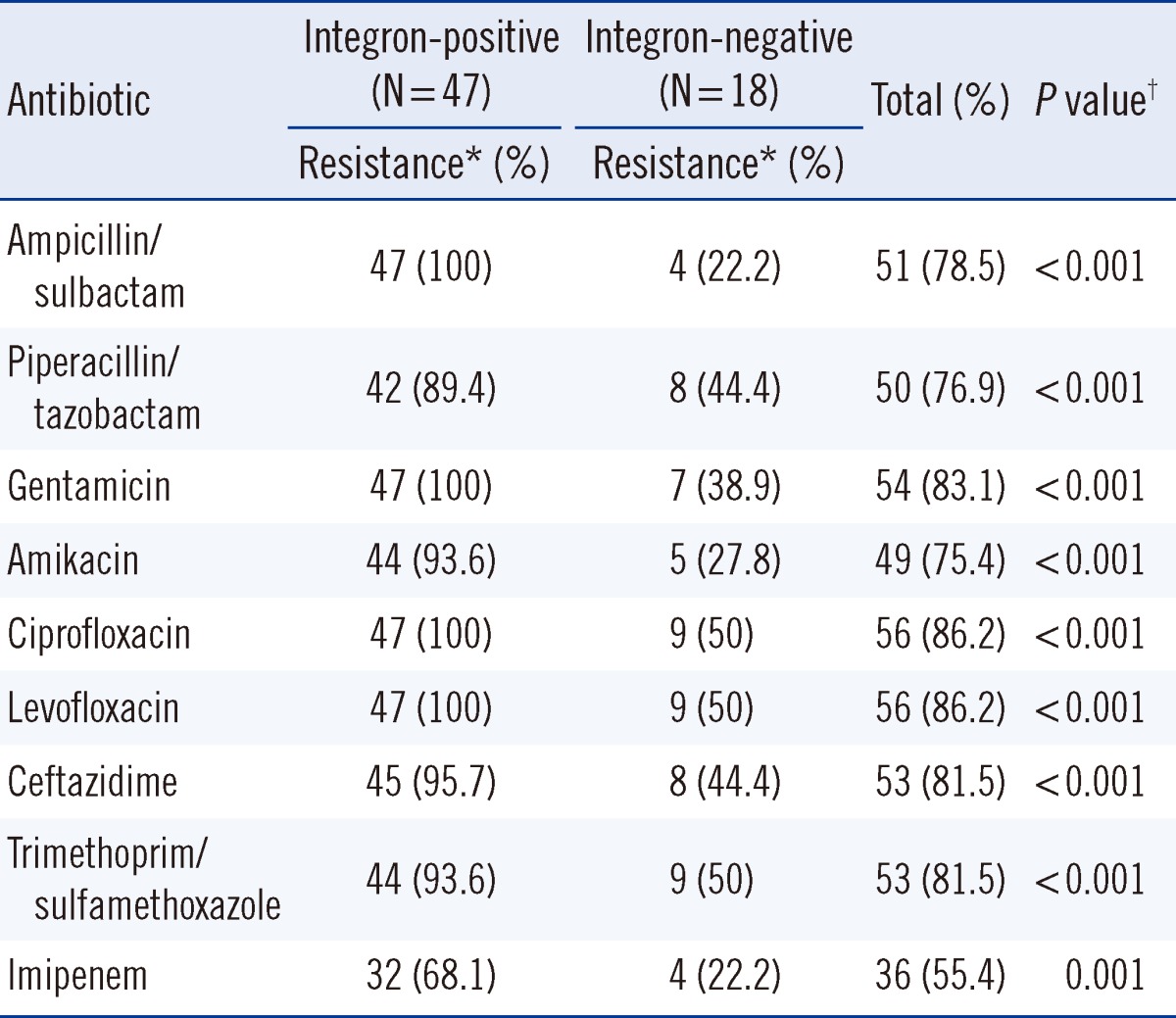
Eighty non-duplicated Acinetobacter species isolates were collected from four regional hospitals in northern Taiwan. Of all the isolates, 65 were identified as A. baumannii, according to the presence of an internal 208-bp fragment from the intergenic spacer region, and the remaining 15 isolates as other Acinetobacter species. Around 72% of the A. baumannii isolates (47/65) carried class 1 integrase genes; however, none of the 65 isolates possessed class 2 integrase genes. The integron-positive strains were significantly more resistant to all of the tested antibiotics compared to the integrase-negative strains (Table 1). In the integron-positive group, the resistance ratios of all the tested antimicrobials, except imipenem, were >89%, while 68% of the isolates were resistant to imipenem.
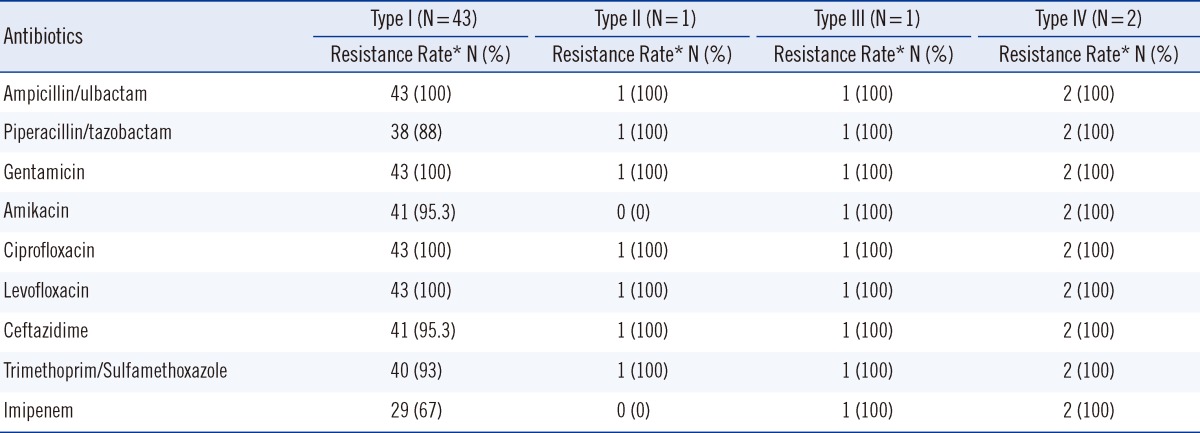
Integron cassette genes were found in all class 1 integrase-containing isolates. Three different gene cassette array fragments, namely, 2.3 kb, 1.8 kb, and 3.0 kb, were detected. They were identified as aacA4-catB8-aadA1 (Genbank accession no. AY557339), dhfrXII-orfF-aadA2 (GenBank accession no. DQ141318), and aacC1-orfP-orfP-orfQ-aadA1 (Genbank accession no. AY577724) by RFLP and sequencing. With the proper restriction enzymes, the amplicons of the three gene cassettes can be divided into two corresponding restriction fragment sizes, which are shown in Table 2. The integrase-containing isolates can further be categorized into four types according to the combination of gene cassette arrays: type I (91.5%, 43/47) containing the 2.3 kb fragment only, type II containing the 2.3 kb and 3.0 kb fragments (2.1%, 1/47), type III containing the 2.3 kb and 1.8 kb fragments (2.1%, 1/47), and type IV containing the 2.3 kb, 3.0 kb, and 1.8 kb fragments (4.3%, 2/47). Table 3 shows the antibiotic resistance phenotypes of the four types of A. baumannii with different gene cassette arrays. All the four types of A. baumannii were multidrug resistant in nature and not susceptible to ampicillin/sulbactam, gentamicin, ciprofloxacin, or levofloxacin. With the exception of type II, 95% of the other types were resistant to amikacin. Despite this, neither the metallo-β-lactamase (MBL) nor the carbapenem-hydrolyzing class D β-lactamase (CHDL) gene-carrying cassettes were found in the integrons. Type I had high ceftazidime (95.3%) and imipenem (67%) resistance rates.
A dendrogram was created to represent the rep-PCR fingerprints of the 38 imipenem-resistant A. baumannii strains (Fig. 1). Of these strains, 84% (32/38) constitutes cluster I and 16% (6/38) constitutes cluster II at an 80% similarity cut-off value.
Integrase gene PCR has been proposed as a method for the routine screening and identification necessary for the surveillance of clinical isolates of A. baumannii with epidemic potential [14]. Previous studies showed that class 1 integrons could be found in 44-84% of A. baumannii clinical isolates [15, 16]. Even in Acinetobacter complex isolates, integron-positive rates can go above 94% [17]. With 72% of the isolates studied in this study carrying integrons and integron-positive strains being more resistant to all of the tested antibiotics, we propose that the presence of integrons can be used as a representative marker of multidrug resistance in A. baumannii.
With regard to genotyping for A. baumannii, pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), and rep-PCR have been used in previous studies [18-20]. While PFGE is still considered the gold standard for typing the outbreak-related isolates of A. baumannii [18], MLST is primarily useful for population genetic studies [19]. Despite inter-laboratory variability of rep-PCR, this method has the advantage of being less labor-intensive than PFGE and MLST. Intra-laboratory clustering of A. baumannii has been proved to be well conserved [20] and to correlate well with MLST results [21], demonstrating the robustness of rep-PCR. In this study, one rep-PCR major cluster (84%) was found among the four regional hospitals in northern Taiwan. It seems that the strains of this cluster spread among these hospitals and disseminated the class I integron with their gene cassettes.
Two of the three gene cassette arrays in the present study, namely, aacA4-catB8-aadA1 and aacC1-orfP-orfP-orfQ-aadA1, have been documented in our previous study [22]. The former cassette array, which can be found in A. baumannii worldwide [15, 23], was the most prevalent (100%) in this study, while the latter was present in only three isolates despite its wide distribution in the strains of the European clones I and II from many countries [24]. Another cassette array dhfrXII-orfF-aadA2, which has been reported in other studies in Taiwan [25], can be found in three isolates. Although there are differences of cassette array types among our study and the previous studies in Taiwan [25, 26], a high percentage of aacA4-catB8-aadA1 existence in class 1 integrons implicates this cassette as a prevalent one and raises the possibility of the same gene cassette presence among epidemiologically unrelated A. baumannii strains.
There was a significant increase in the proportion of the number of health-associated infections caused by carbapenem-resistant A. baumannii over that by all strains of A. baumannii from 14% in 2003 to 46% in 2008 in Taiwan [27]. We found that imipenem-resistant A. baumannii had high integron-carrying rates, although, no MBL or CHDL genes in the integrons were detected in this study. Our previous study disclosed the presence of CHDL in the clinical isolates of A. baumannii from northern Taiwan [28]. However, we did not explore the full spectrum of resistance genes in our clinical isolates any further because we chose to focus on the relationship between the antimicrobial resistance pattern and the gene cassette arrays of integrons in this study. The resistance rates to all the tested antibiotics, except imipenem, of the collected A. baumannii isolates were >70%. However, the presence of integrons with related gene cassettes cannot be responsible for all these resistance phenotypes. Only three categories of gene cassettes, including aminoglycoside-resistant (aaCA4, aacC1, aadA1, aadA2), chloramphenicol-resistant (catB8), and trimethoprim/sulfamethoxazole-resistant (dhfrXII), were identified in spite of previous studies reporting integrons carrying carbepenemase genes.
The whole genome pyrosequencing of an epidemic multidrug-resistant A. baumannii strain belonging to the European clone II group showed the presence of an island containing seven resistance genes, including a class 1 integron carrying the aacA4 and the blaOXA-20 β-lactamase genes [29]. The correlation between the presence of integrons and certain resistance determinants might explain such multidrug resistance manifestation of integron-positive A. baumannii strains. Of the 47 integron-carrying A. baumannii isolates, three harboring aminoglycoside encoding genes did not confer resistance to amikacin. A similar finding was reported by Huang et al. [7]. Further studies are needed to determine whether mutations are occurring inside the gene cassettes or in the upstream sequence of the cassette in order to affect cassette gene expression.
In conclusion, the present work gave a brief description on the molecular epidemiology of A. baumannii integrons and their associated cassette arrays in northern Taiwan. The relationship between antimicrobial resistance and gene cassettes provides insight into the role of integrons in the multidrug resistance of A. baumannii.
Acknowledgements
This study was supported by a grant from the North Region Alliance Department of Health Hospital in Taiwan. We thank Tao-Yuan and Miao-Li General Hospitals, Department of Health (DOH) for providing the studied Acinetobacter strains. The authors also thank Mr. Yeu-Shiuan Sung for technical support.
References
1. Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007; 51:3471–3484. PMID: 17646423.
2. Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008; 21:538–582. PMID: 18625687.
3. Valenzuela JK, Thomas L, Partridge SR, van der Reijden T, Dijkshoorn L, Iredell J. Horizontal gene transfer in a polyclonal outbreak of carbapenem-resistant Acinetobacter baumannii. J Clin Microbiol. 2007; 45:453–460. PMID: 17108068.
4. Carattoli A. Importance of integrons in the diffusion of resistance. Vet Res. 2001; 32:243–259. PMID: 11432416.

5. Mazel D. Integrons: agents of bacterial evolution. Nat Rev Microbiol. 2006; 4:608–620. PMID: 16845431.

6. Gu B, Tong M, Zhao W, Liu G, Ning M, Pan S, et al. Prevalence and characterization of class I integrons among Pseudomonas aeruginosa and Acinetobacter baumannii isolates from patients in Nanjing, China. J Clin Microbiol. 2007; 45:241–243. PMID: 17122024.
7. Huang LY, Chen TL, Lu PL, Tsai CA, Cho WL, Chang FY, et al. Dissemination of multidrug-resistant, class 1 integron-carrying Acinetobacter baumannii isolates in Taiwan. Clin Microbiol Infect. 2008; 14:1010–1019. PMID: 19040472.
8. Turton JF, Kaufmann ME, Glover J, Coelho JM, Warner M, Pike R, et al. Detection and typing of integrons in epidemic strains of Acinetobacter baumannii found in the United Kingdom. J Clin Microbiol. 2005; 43:3074–3082. PMID: 16000417.
9. Chen TL, Siu LK, Wu RC, Shaio MF, Huang LY, Fung CP, et al. Comparison of one-tube multiplex PCR, automated ribotyping and intergenic spacer (ITS) sequencing for rapid identification of Acinetobacter baumannii. Clin Microbiol Infect. 2007; 13:801–806. PMID: 17488329.
10. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk Susceptibility Tests. Approved Standards-Tenth edition, M02-A10. Wayne, PA: Clinical and Laboratory Standards Institute;2009.
11. Koeleman JG, Stoof J, Van Der Bijl MW, Vandenbroucke-Grauls CM, Savelkoul PH. Identification of epidemic strains of Acinetobacter baumannii by integrase gene PCR. J Clin Microbiol. 2001; 39:8–13. PMID: 11136740.
12. Vincze T, Posfai J, Roberts RJ. NEBcutter: a program to cleave DNA with restriction enzymes. Nucleic Acids Res. 2003; 31:3688–3691. PMID: 12824395.

13. Misbah S, AbuBakar S, Hassan H, Hanifah YA, Yusof MY. Antibiotic susceptibility and REP-PCR fingerprints of Acinetobacter spp. isolated from a hospital ten years apart. J Hosp Infect. 2004; 58:254–261. PMID: 15564001.
14. Gaur A, Prakash P, Anupurba S, Mohapatra TM. Possible role of integrase gene polymerase chain reaction as an epidemiological marker: study of multidrug-resistant Acinetobacter baumannii isolated from nosocomial infections. Int J Antimicrob Agents. 2007; 29:446–450. PMID: 17270402.
15. Gallego L, Towner KJ. Carriage of class 1 integrons and antibiotic resistance in clinical isolates of Acinetobacter baumannii from northern Spain. J Med Microbiol. 2001; 50:71–77. PMID: 11192508.
16. Gombac F, Riccio ML, Rossolini GM, Lagatolla C, Tonin E, Monti-Bragadin C, et al. Molecular characterization of integrons in epidemiologically unrelated clinical isolates of Acinetobacter baumannii from Italian hospitals reveals a limited diversity of gene cassette arrays. Antimicrob Agents Chemother. 2002; 46:3665–3668. PMID: 12384388.
17. Sung JY, Kwon KC, Cho HH, Koo SH. Antimicrobial resistance determinants in imipenem-nonsusceptible Acinetobacter calcoaceticus-baumannii complex isolated in Daejeon, Korea. Korean J Lab Med. 2011; 31:265–270. PMID: 22016680.
18. Seifert H, Dolzani L, Bressan R, van der Reijden T, van Strijen B, Stefanik D, et al. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J Clin Microbiol. 2005; 43:4328–4335. PMID: 16145073.
19. Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodríguez-Valera F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol. 2005; 43:4382–4390. PMID: 16145081.
20. Higgins PG, Hujer AM, Hujer KM, Bonomo RA, Seifert H. Interlaboratory reproducibility of DiversiLab rep-PCR typing and clustering of Acinetobacter baumannii isolates. J Med Microbiol. 2012; 61:137–141. PMID: 21903821.
21. Higgins PG, Janssen K, Fresen MM, Wisplinghoff H, Seifert H. Molecular epidemiology of Acinetobacter baumannii bloodstream isolates obtained in the United States from 1995 to 2004 using rep-PCR and multilocus sequence typing. J Clin Microbiol. 2012; 50:3493–3500. PMID: 22895032.
22. Lin MF, Chang KC, Yang CY, Yang CM, Xiao CC, Kuo HY, et al. Role of integrons in antimicrobial susceptibility patterns of Acinetobacter baumannii. Jpn J Infect Dis. 2010; 63:440–443. PMID: 21099097.
23. Zhong Q, Xu W, Wu Y, Xu H. Clonal spread of carbapenem non-susceptible Acinetobacter baumannii in an intensive care unit in a teaching hospital in China. Ann Lab Med. 2012; 32:413–419. PMID: 23130340.
24. Nemec A, Dolzani L, Brisse S, van den Broek P, Dijkshoorn L. Diversity of aminoglycoside-resistance genes and their association with class 1 integrons among strains of pan-European Acinetobacter baumannii clones. J Med Microbiol. 2004; 53:1233–1240. PMID: 15585503.
25. Lee YT, Huang LY, Chen TL, Siu LK, Fung CP, Cho WL, et al. Gene cassette arrays, antibiotic susceptibilities, and clinical characteristics of Acinetobacter baumannii bacteremic strains harboring class 1 integrons. J Microbiol Immunol Infect. 2009; 42:210–219. PMID: 19812854.
26. Su CH, Wang JT, Hsiung CA, Chien LJ, Chi CL, Yu HT, et al. Increase of carbapenem-resistant Acinetobacter baumannii infection in acute care hospitals in Taiwan: association with hospital antimicrobial usage. PLoS One. 2012; 7:e37788. PMID: 22629456.
27. Chen YS, Lin HH, Wu CH, Hsiao YS, Hsu NS, Chen YL. Colonization of a medical center in Southern Taiwan by epidemic strains of carbapenem- and multidrug-resistant Acinetobacter baumannii and the genetic organization of their integrons. Jpn J Infect Dis. 2009; 62:155–157. PMID: 19305060.
28. Lin MF, Chang KC, Lan CY, Chou JL, Kuo JW, Chang CK, et al. Molecular epidemiology and antimicrobial resistance determinants of multidrug-resistant Acinetobacter baumannii among five proximal hospitals in Taiwan. Jpn J Infect Dis. 2011; 64:222–227. PMID: 21617307.
29. Iacono M, Villa L, Fortini D, Bordoni R, Imperi F, Bonnal RJ, et al. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob Agents Chemother. 2008; 52:2616–2625. PMID: 18411315.
Fig. 1
Dendrogram of 38 imipenem-resistant Acinetobacter baumannnii isolates and A. baumannii ATCC19606. 84% (32/38) constitutes cluster I and 16% (6/38) constitutes cluster II at a similarity cut-off value of 80%. The integrase-containing isolates can further be categorized into type I-IV according to the combination of gene cassette arrays.
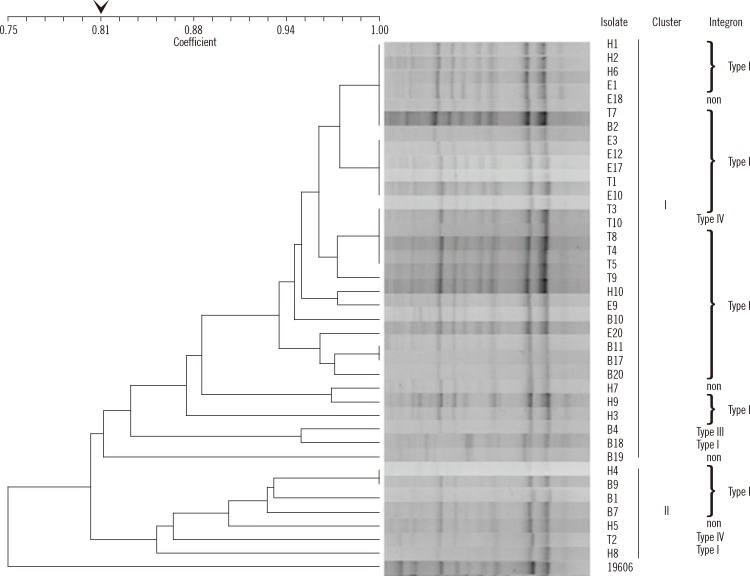
Table 1
Susceptibility testing results of 65 integron-positive and integron-negative Acinetobacter baumannii isolates




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download