Abstract
Objectives
Infants with slight/mild or late-onset hearing impairment might be missed in universal newborn hearing screening (UNHS). We identified the mutation hot spot of common deaf gene in the newborns in Jinan area population by screening the mutation spot with neonate cord blood, in order to make clear whether the neonate cord blood for screening is feasible.
Methods
Six hundred and forty-six newborns were subjected to both UNHS and genetic screening for deafness by using neonate cord blood. The newborn genetic screening targeted four deafness-associated genes, which were commonly found in the Chinese population including gap junction beta-2 protein (GJB2), gap junction beta-3 protein (GJB3), solute carrier family 26 member 4 (SLC26A4), and mtDNA 12S rRNA. The most common 20 spot mutations in 4 deaf genes were detected by MassARRAY iPLEX platform and mitochondrial 12S rRNA A1555G and C1494T mutations were sequenced using Sanger sequencing.
Results
Among the 646 newborns, 635 cases passed the UNHS and the other 11 cases (1.7%) did not. Of the 11 failures, two cases were found to carry homozygous GJB2 p.R143W pathogenic mutation, one case was found to have heterozygous GJB2 235delC mutation, and another one case carried heterozygous GJB3 p.R180X pathogenic mutation. Six hundred and thirty-five babies passed the newborn hearing screening, in which 25 babies were identified to carry pathogenic mutations, including 12 heterozygotes (1.9%) for GJB2 235delC, eight heterozygotes (1.3%) for SLC26A4 IVS7-2A>G, one heterozygote (0.2%) for p.R409H, two homozygotes (0.3%) for m.1494C>T, and two homozygotes (0.3%) for m.1555A>G.
Hearing loss is one of the most common disorders of sensorineural function and it affects at least 30% of the population at some time in their lives. Congenital deafness is one of the common birth defects in human. Approximately 1 out of every 700-1,000 neonates was born with congenital deafness. In terms of the etiology of childhood hearing loss, half of the cases have a genetic cause and 70% of all cases of genetic hearing impairment are nonsyndromal. The most common known cause is autosomal recessive nonsyndromic hearing loss (NSHL). In the Chinese population, studies have shown that a large proportion of nonsyndromic forms of hearing loss is caused by a limited number of genes with recurrent mutations, such as gap junction beta-2 protein (GJB2), gap junction beta-3 protein (GJB3), solute carrier family 26 member 4 (SLC26A4), and mitochondrial DNA (mtDNA) 12S rRNA [1], which would facilitate clinical testing. GJB2 gene mutation can cause autosomal dominant nonsyndromic deafness (DFNA3A) and autosomal recessive nonsyndromic hearing loss (DFNB1A). Forty to sixty percent of patients with sensorineural deafness in early childhood are caused by the GJB2 type of mutation [2], which are characterized as congenital sensorineural hearing loss, noncongenital prelingual deafness and postlingual deafness, with onset age from 6-8 months to 20 years [3]. Carrier frequency of GJB2 gene mutations c.235delC is the most common type of mutational spot in the Chinese population, which can cause severe or profound hearing loss by its homozygous mutations. A mutation in the SLC26A4 gene is the second major contributor to genetic hearing loss, after GJB2 mutations, and is responsible for DFNB4 and enlarged vestibular aqueduct (EVA). Its most common mutational sites is IVS7-2A>G, followed by p.H723R [4]. GJB3 is one of the members of gap junction proteins. GJB3 may cause DFNA2B and DFNB91. m.1494C>T and m.1555A>G in mitochondrial 12S rRNA have close relationship with aminoglycoside antibiotic induced deafness (AAID), both incidences of which increase with age [5].
The incidence of neonates with permanent hearing loss has reached the 1.33-1.86 per thousand, with the implementation of the newborn hearing screening (NHS) and the project for early diagnosis and early intervention hearing loss. The incidence of permanent hearing loss in children continues to increase as the age became older [6]. The proportion of children with hearing loss is up to 2.7 per thousand in children younger than 5 years old, which reaches 3.5 per thousand in adolescents. The changes of incidence of hearing loss occur closely related to deafness induced genetic factors. As NHS was carried out, the assessment and reflection on its effects performed simultaneously. More and more inherent limitations were noticed. Firstly, permanent hearing loss can be detected easily by NHS, whereas the mild hearing loss cannot [7]; secondly, late-onset deafness or progressive deafness cannot be detected by NHS, since these neonates were born with normal hearing. Some diseases, such as cytomegalovirus infection, Pendred syndrome, nonsyndrome with autosomal recessive inheritance of EVA as well as mitochondria gene (mtDNA) 12S rRNA m.1555A>G and m.1494C>T mutations, may result in late-onset hearing with normal hearing at birth. Previous studies have shown that it is crucial for a child that hearing impairment in early childhood was identified before he/she is less than six months, and thus early intervention could be performed. On the other hand, early detection of infant hearing loss has far-reaching significance for later communication, cognition, mental health, career planning and so on [8]. The aim of current study is to know, whether neonatal screening for deafness through cord blood can make up for the inadequacy of the NHS, the common mutational types in neonatal deafness gene in the population of the local, to develop considerable screening program suitable for this region.
Six hundred and forty-six newborns were recruited in current study. All subjects were born in the obstetrical department at Handan Centre Hospital between November 2010 and October 2011.
This study was conducted in accordance with the declaration of Helsinki. This study was conducted with approval from the Ethics Committee of Handan Centre Hospital. Written informed consent was obtained from all participants' guardians.
All the subjects were examined bedside in maternity ward by transient evoked otoacoustic emission. Hearing screening was performed in the maternity ward and the newborns were tested under quiet natural sleeping conditions using ambient noise <30 dB. Every infant was examined at the third day after birth with Accuscreen-otoacoustic emission instrument (MADSEN AccuScreen, GN Otometrics, Taastrup, Denmark). The left and right ear was randomly examined. The Otoacoustic emission instrument displays "PASS" or "REFER" to show normal or not. Those infants with Monaural or binaural fails will be followed up and come back to hospital for recheck in 42 days after birth. Those babies with two times of "REFER" need to further audiological examination and identification of permanent hearing loss (http://www.cdc.gov/ncbddd/ehdi/ehdi.htm).
Up to 2-mL cord blood from umbilical vein were collected into vacuum blood tube with ethylenediaminetetraacetic acid anticoagulant by the obstetrician before the delivery of the placenta. The blood cells were preserved at -80℃ after separation for later gene screening for deafness. Genomic DNA of all subjects was extracted by using AxyPrep genomic DNA extraction Kit (Axygen Biotechnology Co., Silicon Valley, CA, USA), following the kit instructions in the process of extraction.
MassArray system (Sequenom Inc., San Diego, CA, USA) screening for deafness gene array technology platform was used in current study, which is based on matrix assisted laser desorption ionization time of flight mass spectrometry, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) technology (Fig. 1). Assay Designer package (Sequenom Inc.) was used to design polymerase chain reaction (PCR) primers and single base extension primers. 20 sites can be tested simultaneously in one well (Table 1).
PCR amplification of the MassArray system and single base extension were performed strictly accordance with the product instructions. After the multiplex PCR reaction, extra nucleotides were digested by shrimp alkaline phosphatase, and then extended by single base primer. The products were spotted in 384 holes on genotyping chip, and then were analyzed with MALDI-TOF-MS. Final results were read by the MassARRAY RT real-time software systems. Genotype analysis was completed by the MassARRAY Typer software.
3730xl DNA analyzer (Applied Biosystems Inc., Carlsbad, CA, USA) was used for sequencing. The m.1494C>T and m.1555A>G mutational sites in mtDNA 12Sr RNA were screened by using direct sequencing screening, with the primers designed by the online software Primer 3 (http://biotools.umassmed.edu/bioapps/primer3_www.cgi). Forward primer sequences: 5'-CAACCTCACCACCTCTTGCT-3'; reverse primer sequences: 5'-GTAAGGTGGAGTGGGTTTGG-3'. PCR reaction conditions: prede-generated at 94℃ for 30 seconds; annealed at 60℃ for 30 seconds, and 72℃ extracted for 30 seconds, 35 loops; after that, extended at 72℃ for 5 minutes, 1.5% agarose gel electrophoresis to observe the PCR products. DNA mutation-positive samples were reverse sequenced for validation.
Six hundred and forty-six newborns were examined by traditional hearing screening. Among them, 103 cases did not pass the initial screening. In the secondary screening at the 42nd day after birth, a total of 12 cases failed (Table 2). Five cases (0.7%) failed in unilateral hearing examination. Six cases failed in bilateral hearing screening. A total of fail rate of NHS was 1.7%.
Among the 11 cases who failed the NHS, two cases carrying p.R143W homozygous mutation, one case carrying c.235delC single heterozygous mutations in GJB2 gene (suspicious pathogenic mutations), and one case carrying p.R180X pathogenic homozygous mutation in gene GJB3. No hot spot mutation was found in the other seven cases. After full-length sequencing on above four most common deafness genes (GJB2, GJB3, SLC26A4, and mtDNA 12S rRNA), one cases was found carrying c.299-300delAT single heterozygous mutation in GJB2 gene, another one cases carrying p.V37I pathogenic homozygous mutation. No mutation was found in above four common deafness genes in the remaining 5 cases (detailed gene type; Table 3).
Among the 634 newborns who passed the NHS, many cases were found carrying deafness mutations in alleles. A total of 25 cases (3.9%) were found carrying pathogenic mutations (Table 4). Three hundred and thirty-seven cases (53.2%) were found carrying suspected pathogenic mutations (Table 5). Twelve out of 25 pathogenic mutations occurred in GJB2 allele. By sequencing the coding regions of the GJB2 gene, one p.V37I heterozygous mutation was found in one case, and two polymorphic sites p.V27I was found in two cases, no mutation was found in other 10 cases. High rates of possible pathogenic mutations p.E114G in GJB2 gene were found in 292 cases (45.2%), including 54 homozygous mutations and 238 heterozygous mutations. Since all carriers passed the examination of the NHS, the mutation p.E114G is supposed to be polymorphic sites in this region. On gene GJB3, possible pathogenic heterozygous mutation p.V84I was found in only one case, simultaneously with mitochondrial 827A>G mutation. On gene SLC26A4, single heterozygous mutation IVS7-2A>G was found in 8 cases and p.R409H mutation was found in one case. mtDNA 12S rRNA mutations were found in 48 cases, including 21 cases with m.827A>G mutation, 23 cases with m.1005T>C mutation, two cases with m.1494T>C mutation, and two cases with m.1555A>G mutation, all of which were homoplasmic mutations.
Early diagnosis of neonatal hearing impairment plays a very important role in their early treatment or use of hearing aids, and for a better language development. Current paid a close attention to the common deafness genes in neonates by using the MassArray platform and Sanger sequencing. By traditional NHS, hearing impairment can be found in 1.7% of the subjects, whereas up to 4.5% of the subjects were detected carrying pathogenic deafness mutations by gene screening in current study. By sequencing the four common deafness genes of neonates who failed the NHS, the rate of pathogenic rate increased to 4.7% of the subjects who failed NHS. Audiology diagnosis for those who failed NHS could not be made until the neonate is three months old under current diagnostic mode, while gene screening method provides us the accurate information of deafness gene mutations shortly after birth to allow the intervention and treatment made much earlier.
Mutations of GJB2 gene are considered to be the most common cause of sensorineural hearing loss by far. From Table 3, among the 11 neonates who failed the NHS, two types of homozygous mutations were detected in three neonates including p.R143W (NB47 and NB504, 16.7%) and p.V37I (NB10, 8.3%), one single heterozygous mutation c.235delC was detected in one case (NB106). As we have known, homozygous mutations p.R143W in GJB2 gene can cause recessive nonsyndromic sensorineural deafness [9]. High rate of p.R143W mutation in GJB2 gene (0.3%) in Jinan area indicated that it might be one of the major reasons for neonatal hearing loss in this region. On the other hand, c.235delC single heterozygous mutation was found in 13 neonates. No mutation was found in encoding area by sequencing GJB2 gene. Otherwise, there might be mutations in regulatory region in the genes sequenced or mutations in other genes related to the cause of hearing loss. Among 12 neonates who passed NHS, one neonate was found carrying complex heterozygous mutation p.V37I and c.235delC. The carriers with complex heterozygous mutation p.V37I and c.235delC might be normal without hearing impairment when they are born, whereas they often suffer mild, progressive, or asymmetry deafness later. The neonate with NB10 mutation is abnormal with hearing loss in only one ear. This patient also needs follows up, since carriers with p.V37I mutation commonly characterized by progressive deafness [10]. It was known that p.R180X in GJB3 gene could cause DFNA2B. Single heterozygous mutations in NB121 could cause neonatal deafness, consistent with its dominant characteristics of the deaf. Whether if p.V84I mutation is the cause of dominant deafness is not convinced yet, although it is detected frequently in Chinese, which needs further study [11]. In current study, p.V84I mutation was found in only one newborn with normal NHS result.
The nine neonates with IVS7-2A>G and p.R409H mutations in SLC26A4 gene all passed the NHS, who might be just mutation carriers. These carriers still had the possibility of suffering from EVA and nonsyndrome DFNB4. Previous studies showed that patients with EVA and DFNB4 were commonly born with normal hearing or mild hearing impairment, although they got worse at growth stage [12]. From an English study enrolled with 43 cases with EVAs, biallelic gene mutations were found in 17 cases, single allele mutations were found in 16 cases [13]. Generally, double-allele mutations can lead to phenotypic abnormality, however, SLC26A4 gene mutations were found in quite a few subjects with NSHL, what role does single-allele mutations play in NSHL is still a question. Some pathogenic mutations might be located in promoter region or splice site, which were not detected in this study. The current study shows carriers with single-allele gene mutation has more probability to contract EVAS [14]. Those nine newborns with IVS7-2A>G and p.R409H mutations in SLC26A4 gene should be regular followed up and avoid activity that may cause damage to the ears.
mtDNA 12S rRNA m.1494C>T and m.1555A>G were the most two common mutation sites associated with AAID. The detection rates of m.1555A>G mutation (3.96%) was much higher than that of m.1494C>T mutation (0.18%) in AAID patients [15]. The detection rate of m.1555A>G in previous neonatal screening for deafness genes was 0%-0.16% [1617]. In this study, the mutation rate for both m.1555A>G and m.1494C>T were 0.3%, indicated that the detection rates of these two mutations were accordant with each other. It is necessary to incorporate routine screening project. Because mitochondrial genetic are maternally inherited, four cases with positive mutations consisted of three girls and one boy. The study on AAID patients indicated that, the time of aminoglycoside drugs therapy for carriers with m.1494C>T and m.1555A>G mutations was related with their degree of deafness. Aminoglycosides can cause severe or profound deafness before 10 years old [18]. Carriers with m.1494C>T and m.1555A>G mutations might contract late-onset hearing loss with morphological features even without exposure to aminoglycoside antibiotics [1920]. So those four newborns with positive mutations should avoid use of aminoglycoside antibiotics to avoid or postpone hearing loss. The mutation rates of two possible pathogenic mutations m.827A>G and m.1005T>C in mtDNA 12S rRNA was up to 3.3% and 3.6% respectively. The mutation rate of m.827A>G in current study is much higher than that in which the m.827A>G mutation rates are far higher than Brazil neonatal population (0.03%) [16]. The carrier rate of m.1005T>C in 1642 children of Chinese Han people with nonsyndromic deafness was 0.61% [15]. None was reported about the carrier rate in the neonatal population. m.827A>G and m.1005T>C has been reported in AAID pedigrees from various ethnic groups, which showed that above mutations may not cause deafness, but plays an important role in the pathogenesis of AAID. Carriers with above two mutations also need to be informed to avoid the use of aminoglycoside antibiotic and regular follow-up. Carriers with four pathogenic mutations will be studied by family survey.
Some common deafness-associated mutations are associated with mild and/or progressive hearing loss. There will be false-negative results in some neonates with late-onset or mild hearing loss merely by NHS. A retrospective study on probands with deafness found that, up to 3.8%-8% of neonates who passed NHS contracted hearing loss caused by the mutation of GJB2 gene [2122]. Young et al. [23] reported that, among 108 cases who received the treatment of cochlear implantation, 32 cases (29.6%) had passed the examination of NHS before eight years. Which indicated there is a limitation of the NHS in recognition of patients with late-onset deafness. How to identify late-onset or progressing deafness in the baby or child earlier have been in the project of Early Hearing Detection and Intervention (http://www.CDC.gov/NCBDDD/EHDI/nationalgoals.htm), but there are not very effective method to achieve this goal. Some common GJB2 mutations, such as p.M34T and p.V37I are associated with mild-to-moderate SNHI [24]. Previous studies indicated that mutation screening for GJB2 deafness gene is valuable for judging, closely observed, and early detected for the baby with progressive or late-onset deafness with practicality, making these babies were closely observed, progress in the early detection of their deafness. Detection of m.1494C>T and m.1555A>G mutations in SLC26A4 gene and mtDNA 12S rRNA was of greater significance for the outcome. For carriers with m.1494C>T and m.1555A>G mutations in mtDNA 12S rRNA, deafness causation is the greatest attraction of aminoglycoside [18], so such mutations carriers you want to absolutely avoid the use of aminoglycoside antibiotics. Aminoglycoside antibiotics widely used in China, the latest survey found that in 28% of 1,642 patients with whole hearing loss were induced by aminoglycoside drugs [15]. In other words, newborn genetic screening might be useful for identifying slight/mild hearing loss that was not detected by conventional NHS.
The project of implementation of neonatal screening for deafness genes is different for different neonates. Although the GJB2 gene is considered to be the most common cause of hereditary deafness in most population of the world, the mutations rates are various in different races or in different regions. As to China c.235delC and p.V37I are common mutations [25]. The prevalence of EVA and DFNB4 caused by SLC26A4 gene mutation and nonsyndromic GJB2 gene mutation is the second followed by GJB2 gene mutation [26], whose mutation types also differ in the different populations. p.T416P and c.1001+1G>A are common mutations in Nordic population [27]; p.H723R is the most common form of mutant in Japan [26] and Korea [25] population. The most common mutation in Chinese population is IVS7-2A>G, followed by p.H723R; but in patients with nonsyndromic deafness in Tibetan of China, carrier rate of SLC26A4 mutation in GJB2 gene is much lower than other region [28]. m.1555A>G mutation in mtDNA 12S rRNA in deaf patients was reported all over the world, the mutation rate of m.1494C>T mutation in Chinese patients with different phenotypic of sensorineural deafness is only 0.1%, much lower than that of m.1555A>G (1.72%). In this study, the mutation rate of m.1494C>T and m.1555A>G are the same in newborns (both are 0.3%). Furthermore, besides the common c.235delC in Chinese population, the carrier rates of p.R143W is high in Jinan area. IVS7-2A>G is the most common mutation in SLC26A4 gene. It is thus clear that the screening program in different regions should be developed according to genetic testing results for people on the ground.
In short, neonatal screening for common deafness gene, can explores the causation of hearing impairment of newborns who failed the NHS on the molecular level, help us to find patients with mild hearing loss and aminoglycoside antibiotic-sensitive carriers, and provides early guidance to rationality of neonatal medicine, so as to effectively prevent or delay hearing loss. This research got the information on the local carries with mutations in the deafness gene, and laid a good foundation for promoting neonatal screening for deafness gene in the future.
ACKNOWLEDGMENTS
The authors would like to thank the patients who participated in this study and the research teams of the Birth Defect Research Center & Pathology Research Center, Fudan University, Shanghai, China, who assisted with this research. This work was supported by the Key Program of Handan (1113108017) and the Science and Technology Support Program of Hebei (11276102D).
References
1. Ouyang XM, Yan D, Yuan HJ, Pu D, Du LL, Han DY, et al. The genetic bases for non-syndromic hearing loss among Chinese. J Hum Genet. 2009; 3. 54(3):131–140. PMID: 19197336.

2. Propst EJ, Stockley TL, Gordon KA, Harrison RV, Papsin BC. Ethnicity and mutations in GJB2 (connexin 26) and GJB6 (connexin 30) in a multi-cultural Canadian paediatric Cochlear Implant Program. Int J Pediatr Otorhinolaryngol. 2006; 3. 70(3):435–444. PMID: 16125251.

3. Snoeckx RL, Huygen PL, Feldmann D, Marlin S, Denoyelle F, Waligora J, et al. GJB2 mutations and degree of hearing loss: a multicenter study. Am J Hum Genet. 2005; 12. 77(6):945–957. PMID: 16380907.
4. Wang QJ, Zhao YL, Rao SQ, Guo YF, Yuan H, Zong L, et al. A distinct spectrum of SLC26A4 mutations in patients with enlarged vestibular aqueduct in China. Clin Genet. 2007; 9. 72(3):245–254. PMID: 17718863.

5. Kokotas H, Petersen MB, Willems PJ. Mitochondrial deafness. Clin Genet. 2007; 5. 71(5):379–391. PMID: 17489842.

6. Kennedy C, McCann D. Universal neonatal hearing screening moving from evidence to practice. Arch Dis Child Fetal Neonatal Ed. 2004; 9. 89(5):F378–F383. PMID: 15321952.

7. Johnson JL, White KR, Widen JE, Gravel JS, James M, Kennalley T, et al. A multicenter evaluation of how many infants with permanent hearing loss pass a two-stage otoacoustic emissions/automated auditory brainstem response newborn hearing screening protocol. Pediatrics. 2005; 9. 116(3):663–672. PMID: 16140706.

8. Yoshinaga-Itano C, Sedey AL, Coulter DK, Mehl AL. Language of early- and later-identified children with hearing loss. Pediatrics. 1998; 11. 102(5):1161–1171. PMID: 9794949.

9. Brobby GW, Muller-Myhsok B, Horstmann RD. Connexin 26 R143W mutation associated with recessive nonsyndromic sensorineural deafness in Africa. N Engl J Med. 1998; 2. 338(8):548–550. PMID: 9471561.

10. Li L, Lu J, Tao Z, Huang Q, Chai Y, Li X, et al. The p.V37I exclusive genotype of GJB2: a genetic risk-indicator of postnatal permanent childhood hearing impairment. PLoS One. 2012; 7(5):e36621. PMID: 22574200.

11. Oh SK, Choi SY, Yu SH, Lee KY, Hong JH, Hur SW, et al. Evaluation of the pathogenicity of GJB3 and GJB6 variants associated with nonsyndromic hearing loss. Biochim Biophys Acta. 2013; 1. 1832(1):285–291. PMID: 22617145.

12. Jackler RK, De La Cruz A. The large vestibular aqueduct syndrome. Laryngoscope. 1989; 12. 99(12):1238–1242. PMID: 2601537.

13. Reardon W, OMahoney CF, Trembath R, Jan H, Phelps PD. Enlarged vestibular aqueduct: a radiological marker of pendred syndrome, and mutation of the PDS gene. QJM. 2000; 2. 93(2):99–104. PMID: 10700480.

14. Pryor SP, Madeo AC, Reynolds JC, Sarlis NJ, Arnos KS, Nance WE, et al. SLC26A4/PDS genotype-phenotype correlation in hearing loss with enlargement of the vestibular aqueduct (EVA): evidence that Pendred syndrome and non-syndromic EVA are distinct clinical and genetic entities. J Med Genet. 2005; 2. 42(2):159–165. PMID: 15689455.

15. Lu J, Li Z, Zhu Y, Yang A, Li R, Zheng J, et al. Mitochondrial 12S rRNA variants in 1642 Han Chinese pediatric subjects with aminoglycoside-induced and nonsyndromic hearing loss. Mitochondrion. 2010; 6. 10(4):380–390. PMID: 20100600.

16. Nivoloni Kde A, da Silva-Costa SM, Pomílio MC, Pereira T, Lopes Kde C, de Moraes VC, et al. Newborn hearing screening and genetic testing in 8974 Brazilian neonates. Int J Pediatr Otorhinolaryngol. 2010; 8. 74(8):926–929. PMID: 20538352.
17. Wu CC, Hung CC, Lin SY, Hsieh WS, Tsao PN, Lee CN, et al. Newborn genetic screening for hearing impairment: a preliminary study at a tertiary center. PLoS One. 2011; 6(7):e22314. PMID: 21811586.

18. Guan MX. Mitochondrial 12S rRNA mutations associated with aminoglycoside ototoxicity. Mitochondrion. 2011; 3. 11(2):237–245. PMID: 21047563.

19. Zhao H, Li R, Wang Q, Yan Q, Deng JH, Han D, et al. Maternally inherited aminoglycoside-induced and nonsyndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese family. Am J Hum Genet. 2004; 1. 74(1):139–152. PMID: 14681830.

20. Lu J, Qian Y, Li Z, Yang A, Zhu Y, Li R, et al. Mitochondrial haplotypes may modulate the phenotypic manifestation of the deafness-associated 12S rRNA 1555A>G mutation. Mitochondrion. 2010; 1. 10(1):69–81. PMID: 19818876.
21. Schimmenti LA, Martinez A, Telatar M, Lai CH, Shapiro N, Fox M, et al. Infant hearing loss and connexin testing in a diverse population. Genet Med. 2008; 7. 10(7):517–524. PMID: 18580690.

22. Norris VW, Arnos KS, Hanks WD, Xia X, Nance WE, Pandya A. Does universal newborn hearing screening identify all children with GJB2 (Connexin 26) deafness? Penetrance of GJB2 deafness. Ear Hear. 2006; 12. 27(6):732–741. PMID: 17086082.

23. Young NM, Reilly BK, Burke L. Limitations of universal newborn hearing screening in early identification of pediatric cochlear implant candidates. Arch Otolaryngol Head Neck Surg. 2011; 3. 137(3):230–234. PMID: 21422305.

24. Pollak A, Skorka A, Mueller-Malesińska M, Kostrzewa G, Kisiel B, Waligora J, et al. M34T and V37I mutations in GJB2 associated hearing impairment: evidence for pathogenicity and reduced penetrance. Am J Med Genet A. 2007; 11. 143A(21):2534–2543. PMID: 17935238.
25. Park HJ, Shaukat S, Liu XZ, Hahn SH, Naz S, Ghosh M, et al. Origins and frequencies of SLC26A4 (PDS) mutations in east and south Asians: global implications for the epidemiology of deafness. J Med Genet. 2003; 4. 40(4):242–248. PMID: 12676893.

26. Miyagawa M, Nishio SY, Usami S. Deafness Gene Study Consortium. Mutation spectrum and genotype-phenotype correlation of hearing loss patients caused by SLC26A4 mutations in the Japanese: a large cohort study. J Hum Genet. 2014; 5. 59(5):262–268. PMID: 24599119.

27. Choi BY, Stewart AK, Madeo AC, Pryor SP, Lenhard S, Kittles R, et al. Hypo-functional SLC26A4 variants associated with nonsyndromic hearing loss and enlargement of the vestibular aqueduct: genotype-phenotype correlation or coincidental polymorphisms? Hum Mutat. 2009; 4. 30(4):599–608. PMID: 19204907.
28. Yuan Y, Zhang X, Huang S, Zuo L, Zhang G, Song Y, et al. Common molecular etiologies are rare in nonsyndromic Tibetan Chinese patients with hearing impairment. PLoS One. 2012; 7(2):e30720. PMID: 22389666.

Fig. 1
Genotype mass spectra of mutational sites. (A) GJB2 c.235delC heterozygous mutation; (B) GJB2 p.R143W homozygous mutation; (C) GJB3 p.V84I heterozygous mutation; (D) GJB3 p.R180X heterozygous mutation; (E) SLC26A4 IVS7-2A>G heterozygous mutation; (F) SLC26A4 p.R409H heterozygous mutation. GJB2, gap junction beta-2 protein; GJB3, gap junction beta-3 protein; SLC26A4, solute carrier family 26 member 4. a, wild type; b, mutant type.
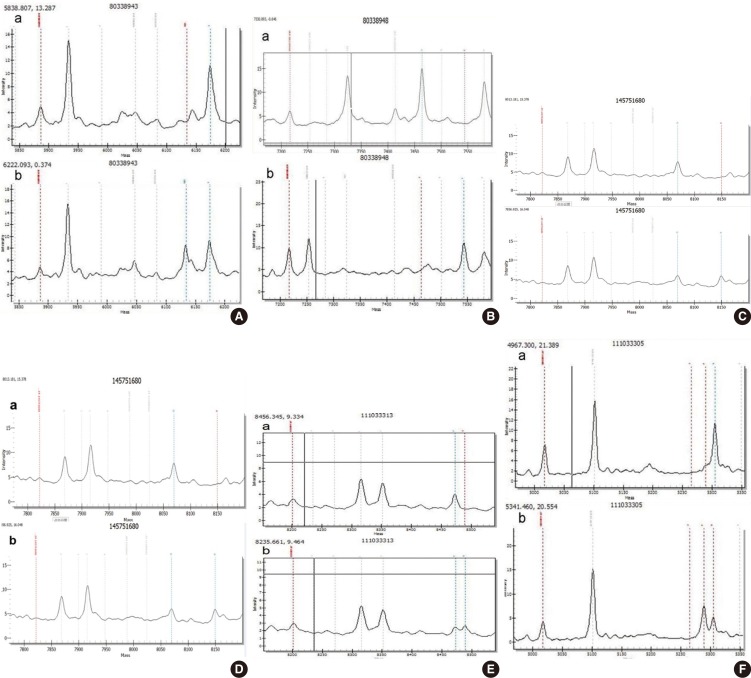
Table 1.
The 20 common deafness mutational sites in current screening
Table 2.
Hearing screening results of 646 newborns
| Result of OAE | No. of cases (%) |
|---|---|
| Both ears passed | 635 (98.3) |
| One ear referred | 5 (0.7) |
| Both ears referred | 6 (1.0) |
| Total cases | 646 (100) |
| Total failed in NHS | 11 (1.7) |
Table 3.
Gene screening results of 11 neonates who failed in newborn hearing screening
Table 4.
Gene screening and newborn hearing screening results of neonates with pathogenic mutations
Table 5.
Gene screening and newborn hearing screening results of neonates with suspected pathogenic mutations
| Genotype |
Newborn hearing screening |
Total | |
|---|---|---|---|
| Pass | Refer | ||
| GJB2 | |||
| p.E114G/ p. E114G | 54 | 0 | 54 |
| p. E114G/wt | 238 | 0 | 238 |
| GJB3 | |||
| p.V84I/wt | 1 | 0 | 1 |
| mtDNA 12S rRNA | |||
| m.827A>G | 21 | 0 | 21 |
| m.1005T>C | 23 | 0 | 23 |
| Total | 337 | 0 | 337 |




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download